The melting temperature calculations are based on the simple thermodynamic relationship between entropy, enthalpy, free energy and temperature, where
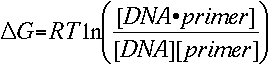
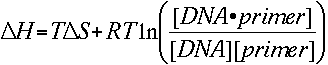
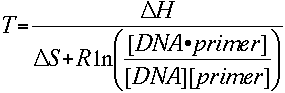
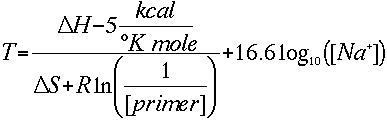
| ASSUMPTIONS: The thermodynamic calculations assume that the annealing occurs at pH 7.0. The melting temperature (Tm) calculations assume the sequences are not symmetric and contain at least one G or C. The oligonucleotide sequence should be at least 8 bases long to give reasonable Tms. |
where w,x,y,z are the number of the bases A,T,G,C in the sequence,
respectively.
| ASSUMPTIONS: Both equations assume that the annealing occurs under the standard conditions of 50 nM primer, 50 mM Na+, and pH 7.0. |
where w,x,y,z are the number of the bases A,T,G,C in the sequence,
respectively.
The following equation is provided only for your reference. It is not actually used by OligoCalc. It is reportedly more accurate for longer sequences.
For more information please see the reference:
Howley, P.M; Israel, M.F.;
Law, M-F.; and M.A. Martin "A rapid method for detecting and mapping homology
between heterologous DNAs. Evaluation of polyomavirus genomes." J. Biol.
Chem. 254, 4876-4883, 1979.
RNA melting temperatures
| ASSUMPTIONS: These equations assume that the annealing occurs under the standard conditions of 50 nM primer and pH 7.0. |
DNA Molecular Weight (for instance synthesized Oligonucleotides)
Anhydrous Molecular Weight = (An x 313.21) + (Tn x 304.2) + (Cn x 289.18) + (Gn x 329.21) - 61.96
An, Tn, Cn, and Gn are the number of each respective nucleotide within the polynucleotide. The subtraction of 61.96 gm/mole from the oligonucleotide molecular weight takes into account the removal of HPO2 (63.98) and the addition of two hydrogens (2.02).
Please note: this calculation works well for synthesized oligonucleotides. If you would like an accurate MW for restriction enzyme cut DNA, please use:
Molecular Weight = (An x 313.21) + (Tn x 304.2) + (Cn x 289.18) + (Gn x 329.21) + 79.0
The addition of 79.0 gm/mole to the oligonucleotide molecular weight takes into account the 5' monophosphate left by most restriction enzymes. No phosphate is present at the 5' end of strands made by primer extension, so no adjustment should be necessary. Also, if you are making these calculations for dsDNA, the correct amount to add it 158.0 gm/mole, adding a monophosphate to each strand.
RNA Molecular Weight (for instance from an RNA transcript)
Molecular
Weight = (An x 329.21) + (Un x 306.17) + (Cn x
305.18) + (Gn x 345.21) + 159.0
An, Un,
Cn, and Gn are the number of each respective nucleotide
within the polynucleotide. Addition of 159.0 gm/mole to the molecular weight
takes into account the 5' triphosphate.
| Residue | Moles-1 cm-1 | Amax(nm) | Molecular Weight (after protecting groups are removed) |
|---|---|---|---|
| Adenine (dAMP, Na salt) | 15200 | 259 | 313.21 |
| Guanine (dGMP, Na salt) | 12010 | 253 | 329.21 |
| Cytosine (dCMP, Na salt) | 7050 | 271 | 289.18 |
| Thymidine (dTMP, Na salt) | 8400 | 267 | 304.2 |
| dUradine (dUMP, Na salt) | 9800 | - | 290.169 |
| dInosine (dUMP, Na salt) | - | - | 314 |
| RNA nucleotides | |||
| Adenine (AMP, Na salt) | 15400 | 259 | 329.21 |
| Guanine (GMP, Na salt) | 13700 | 253 | 345.21 |
| Cytosine (CMP, Na salt) | 9000 | 271 | 305.18 |
| Uradine (UMP, Na salt) | 10000 | 262 | 306.2 |
| Other nucleotides | |||
| 6' FAM | 20960 | 537.46 | |
| TET | 16255 | 675.24 | |
| HEX | 31580 | 744.13 | |
| TAMRA | 31980 | ||
Assume 1 OD of a standard 1ml solution, measured in a cuvette with a 1 cm pathlength.
| Chemical name: | 6-carboxyfluorescein |
| Absorption wavelength maximum: | 495 nm |
| Emission wavelength maximum: | 521 nm |
| Molar Absorptivity at 260nm: | 20960 Moles-1 cm-1 |
| Chemical name: | 4, 7, 2', 7'-Tetrachloro-6-carboxyfluorescein |
| Absorption wavelength maximum: | 519 nm |
| Emission wavelength maximum: | 539 nm |
| Molar Absorptivity at 260nm: | 16255 Moles-1 cm-1 |
| Chemical name: | 4, 7, 2', 4', 5', 7'-Hexachloro-6-carboxyfluorescein |
| Absorption wavelength maximum: | 537 nm |
| Emission wavelength maximum: | 556 nm |
| Molar Absorptivity at 260nm: | 31580 Moles-1 cm-1 |
| Chemical name: | N, N, N', N'-tetramethyl-6-carboxyrhodamine |
| Absorption wavelength maximum: | 555 nm |
| Emission wavelength maximum: | 580 nm |
| Molar Absorptivity at 260nm: | 31980 Moles-1 cm-1 |
| Symbol: nucleotide(s) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
| ||||||||||||||||||||||||||||||||||
Most recent version is available at URL: http://www.basic.northwestern.edu/biotools/oligocalc.html
The current version is the result of efforts by the following people:
Qing Cao, M.S.
Research Computing
Northwestern University Medical
School
Chicago, IL 60611
Warren A. Kibbe, Ph.D.
and PH entry.
Research Computing
Northwestern University
Medical School
Chicago, IL 60611
Original code by Eugen Buehler
Research Support
Facilities
Department of Molecular Genetics and Biochemistry
University of
Pittsburgh School of Medicine
Monomer structures and molecular weights provided by Bob Somers, Ph.D. at Glen Research Corporation
Uppercase/lowercase strand complementation problem described by Alexey Merz
The RNA calculations and functions were requested by Suzanne Kennedy, Ph.D. at Qiagen
The flourescent tags and tagging options were requested by and the data provided by Florian Preitauer and Regina Bichlmaier, Ph.D. at metabion GmbH
The dsDNA and dsRNA molecular weight calculation problems in version 3.xx and fixed in 3.10 was reported by Borries Demeler, Ph.D. and Regina Bichlmaier, Ph.D. at metabion GmbH