Protein Methylation
Proteins are controlled or regulated by a large battery of post-translational modifications (PTMs). Post-translational modification of proteins refers to biochemical events that proteins undergo after translation. Examples of these modification types are methylation, phosphorylation, glycosylation, sulfation, sumoylation, ubiquitinylation, glycosylation, disulfide bonding and others. Post-translational modifications create sites for specific protein-interaction domains. Biochemical research performed during the last decades revealed that in cells protein domains interact in a controlled but dynamic manner and are an important part of regulatory cellular metabolic pathways. Furthermore, post-translational modifications of protein domains define the state of the proteome, a part of cellular organization.
More recently, the inhibition or modulation of protein-protein interactions (PPIs) has become popular in the drug discovery industry. In contrast to traditional drug discovery approaches, the goal is to use the knowledge gained from the study of protein-protein interactions to find new drug candidates or inhibitors. One example for targeted proteins is the protein kinase family. Another example is the search for drugs that allow the treatment of so called “undruggable” diseases.
Methylation alters protein domains or peptide motifs
Proteins can be altered by a diverse set of post-translational modifications. These include the methylation of arginine residues, the methylation, acetylation, ubiquitylation or sumoylation of lysine residues, or prolyl-hydroxylation, and many others. In addition, the presence of multiple post-translational modifications on one protein can have combinatorial effects which can fine tune the regulation of intra-molecular interactions of domain containing proteins.
Today, protein methylation is of wide interest to the scientific field. This hasn’t been always so. Researchers began to study protein methylation in the 1960s, and by the early 1980s, it was known that lysine, arginine, histidine and dicarboxylic amino acids were post-translationally modified. Highly specific enzymes called methyltransferases are known to be responsible for the selective transfer of a methyl group to a targeted molecule. With the availability of modern molecular biology techniques starting in the mid 1990s, is has now become clear that protein methylation is involved in many important functions, including gene regulation and signal transduction.
What is protein methylation?
In chemistry, biochemistry, and biology methylation refers to the addition of methyl groups (-CH3; the addition of 15 mass units or delta mass = 15 dalton) to organic compounds. This reaction type is a specific case of alkylation. The term alkylation refers to the transfer of an alkyl group, such as the isopropyl group, –CH(CH3)2, or the methyl group, -CH3, from one molecule to another. Alkyl groups may be transferred as alkyl groups with a positively charged carbon atom, a free radical, a carbanion or a carbine. In chemistry, some typical methylation reagents are dimethylsulfate, methanol, methyl halides and diazomethane.
However, it is well established now that many different molecules present in a cell can be methylated. For example, scientists now know that methylation of DNA is epigentically inherited. The methylation of DNA turned out to be an important regulator of gene transcription. The addition of methyl groups to DNA typically occurs at CpG islands or regions. In addition, methylation can also occur in RNA molecules, for example at conserved sequence regions in 18S rRNA. Methylation of cytosine residues in DNA to form 5-methylcytosine has widespread effects on gene expression due to recruiting specific DNA-binding proteins.
In cellular proteins many protein or peptide motifs that contain lysine residues can be methylated or acetylated. These modified motifs can lead to the recognition by other protein domains such as chromo-domains or bromo-domains. Such domains are found in proteins regulating chromatin structure and gene expression. For example, the flexible N-terminal and C-terminal ends of histones are known to contain lysine modifications important for the coupling of histones to changes in chromatin organization and the epigenetic control of gene expression. Typically, a single chromo- or bromo-domain recognizes a suitable modified lysine residue within a short peptide motif sequence.
The monomethylation of a lysine side chain is illustrated in Figure 1 and the structure of S-Adenosyl methionine (SAM), the primary methyl donor molecule in cellular metabolism for numerous biochemical reactions is shown in Figure 2
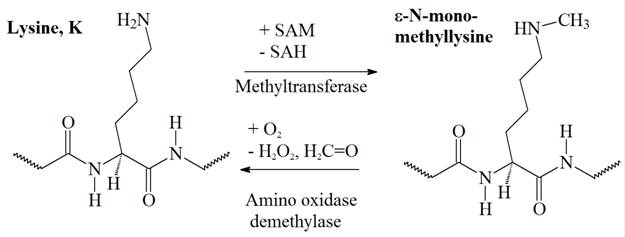

Figure 1: Monomethylation of a lysine on its ε-amino group. Adenosyl methionine (SAM), a molecule composed of adenosine and methionine, is the primary methyl group donor in metabolism. Donation of the methyl group, in this reaction to the ε-amino group of a lysine, transforms SAM into S-adenyl homocysteine (SAH).


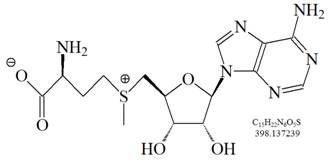
Figure 2: S-Adenosyl methionine (SAM), the primary methyl donor molecule in cellular metabolism for numerous biochemical reactions. During the methionine cycle, methionine is converted to SAM. After transfering its methyl group to a target molecule, SAM is converted to S-adenosyl homocysteine (SAH), which is then further converted to homocysteine. Homocysteine is either converted back to methionine, or enters the trans-sulfuration pathway to form other sulfur-containing amino acids.
The study of lysine methylation of histones has been quite fruitful in recent years after histone lysine methyltransferases were discovered. Histone lysine methylation, either activate or repress gene expression depending on the status of the methylated lysine and its position. However, some of the recently discovered lysine methyl transferases target not only histones but other proteins as well. For example, Set9, a SET domain-containing lysine methyltransferase, was initially found to target histone H3 lysine 4 for mono-methylation but was subsequently shown to target a variety of non-histone proteins as well. Some of its targets are transcription factors.
The SET domain is a 130 to 140 amino acid, evolutionary well conserved sequence motif initially characterized in the Drosophila proteins Su(var)3-9, Enhancer-of-zeste and Trithorax, from which the acronym SET is derived. In addition, the SET domain is found in proteins of diverse functions ranging from yeast to mammals, but also in some bacteria and viruses. Several structures of SET domain proteins have been reported over the past year. SET domains are folded in a novel way, and adjacent domains are used for both structural stabilization and the completion of their active sites. In addition, the cofactor S-adenosyl-L-methionine and peptide substrates bind on opposite faces of the SET domain. Furthermore, the side chain of the target lysine approaches the transferred methyl group through a narrow channel that passes through the middle of the domain.
Reference
Paik WK, Paik DC, Kim S.; Historical review: the field of protein methylation. Trends Biochem Sci. 2007 Mar;32(3):146-52. Epub 2007 Feb 8.
Bruce T. Seet, Ivan Dikic, Ming-Ming Zhou and Tony Pawson; Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006 Jul;7(7):473-83.
SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domain http://smart.embl.de/smart/do_annotation.pl?DOMAIN=SM00317
Xiao B, Wilson JR, Gamblin SJ.; SET domains and histone methylation. Curr Opin Struct Biol. 2003 Dec;13(6):699-705.
Yang XD, Lamb A, Chen LF; Methylation, a new epigenetic mark for protein stability. Epigenetics. 2009 Oct 1;4(7):429-33. Epub 2009 Oct 10.