What are MicroRNAs or miRNAs?
Micro RNAs (miRNAs) are a class of endogenous small non-coding RNAs (ncRNAs) approximately 21 to 24 nucleotides in length found in plants and animals including humans. miRNAs have recently emerged as key regulators in many biological pathways. miRNAs function in the post-transcriptional regulation of gene expression. Processing of miRNAs occurs from approximately 70 nucleotides in size hairpin precursor RNAs by the protein Dicer. miRNA have been shown to regulate their target messenger RNA (mRNA) by destabilizing of mRNA molecules and translational repression. More than half of all messenger RNAs (mRNAs) are estimated to be targets of miRNAs. Each miRNA molecule is predicted to regulate up to hundreds of targets. miRNA appear to regulate a broad range of cellular processes, including proliferation, differentiation, and apoptosis by interacting with specific mRNAs through complementary base-pairing.
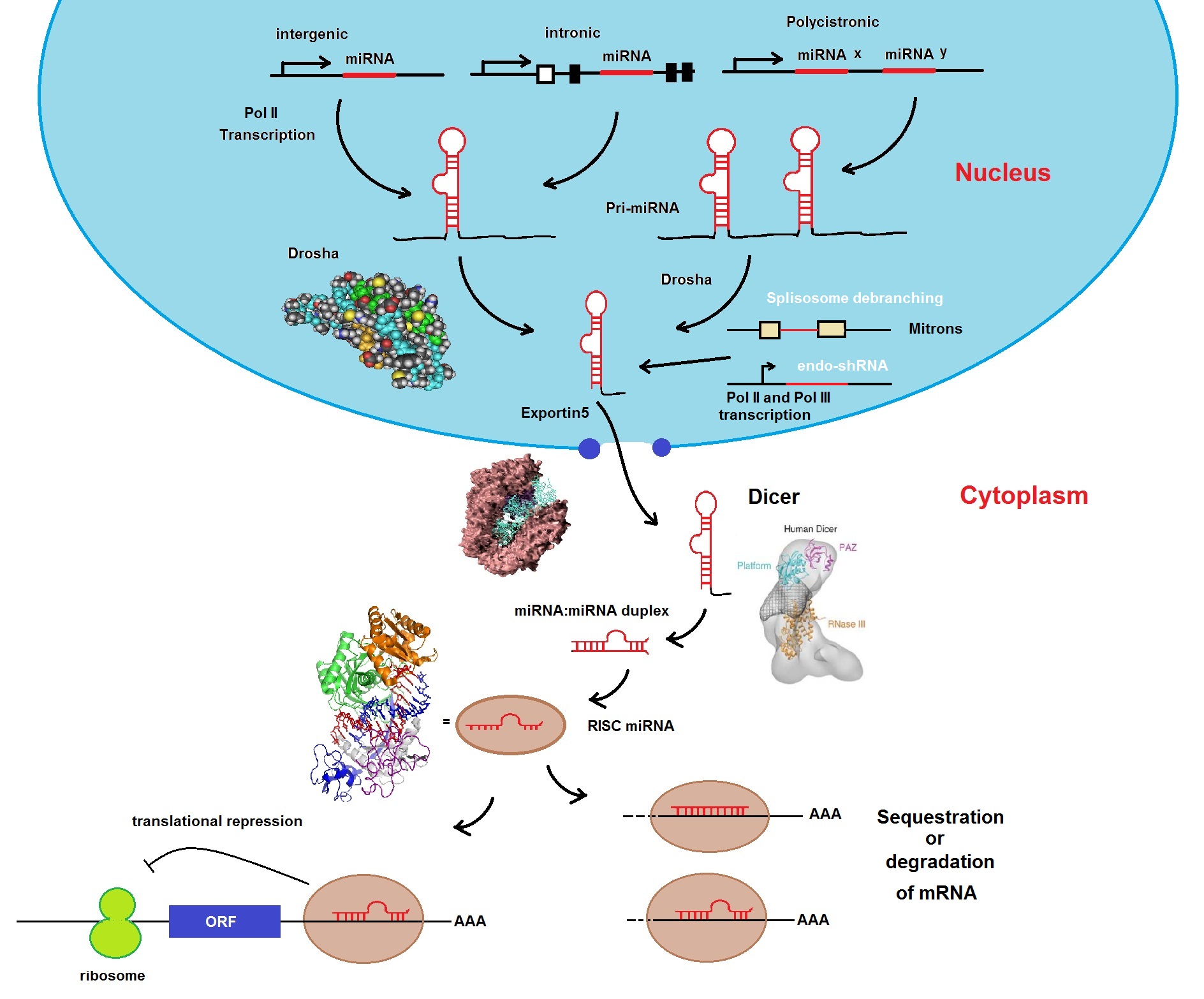
Figure 1: The miRNA biogenesis pathway. miRNAs are transcribed by RNA polymerase II as precursor RNAs from intergenic, intronic or polycistronic genomic loci in combination with specific transcription factors (TF). Following transcription, the transcripts are processed in the nucleus by the RNase III enzyme Drosha in complex with DGCR8 (DiGeorge syndrome chromosomal region 8) into pre-miRNAs. Pre-miRNAs are exported into the cytoplasm by the protein Exportin 5. Pre-miRNAs are further processed by the RNAs III enzyme Dicer together with TRBP (TAR RNA binding protein encoded by the TRBP gene) into a duplex consisting of a guide strand (miRNA) and passenger strand (miRNA*). Next, the mature miRNA is loaded into the RNA-induced silencing complex, or RISC, and acts as a guide strand that based on sequence complementarities recognize target mRNA. RISC inhibits translation or promotes destabilization of target mRNAs.
Large scale production of miRNAs can be achieved by in-vitro transcription (IVT) with the help of T7 RNA polymerase and fully complementary ds DNA oligonucleotides containing a T7 promoter.
The expression of miRNAs is post-transcriptionally regulated by modulation of their maturation. Initially miRNAs are transcribed with much longer RNAs called precursor microRNAs (pri-miRNAs). These are subsequently processed in a series of steps to produce the mature miRNA. First, the long hairpin structure that contains the mature miRNA is cleaved from the primary transcript. The Microprocessor multiprotein complex enables this processing step. The protein complex distinguishes hairpins that contain miRNAs from other hairpin structures in the transcriptome. The excised hairpin is exported to the cytoplasm. In the cytoplasm, the nuclease Dicer cleaves the mature miRNA from the precursor hairpin (pre-miRNA). The pre-element structure is removed. This structure contains a short stem-loop, the terminal loop and a short region of predicted base pairing, also known as the terminal loop region or apical region. The remaining duplex associates with an Argonaute protein. The single-stranded miRNA is then retained in the active miRNA-Argonaute complex.
An understanding of factors that regulate miRNA expression is important for the development of therapeutics that target disease-related miRNAs. To achieve this, Chiravil et al. have solved part of the structure for miR-21 pre-element. Figure 1 shows an NMR based model of a miRNA pre-element, miR-21 pre-element. miR-21 is a miRNA that is elevated in both cancer and heart disease. Furthermore, it is highly expressed in a variety of tumors, contributing to the cancer phenotype by lowering translation of tumor suppressor genes. miR-21 is also expressed in hypertrophic heart tissue, where it contributes to the fibrotic response to cardiac stress or injury (Chirayil et al. 2014).
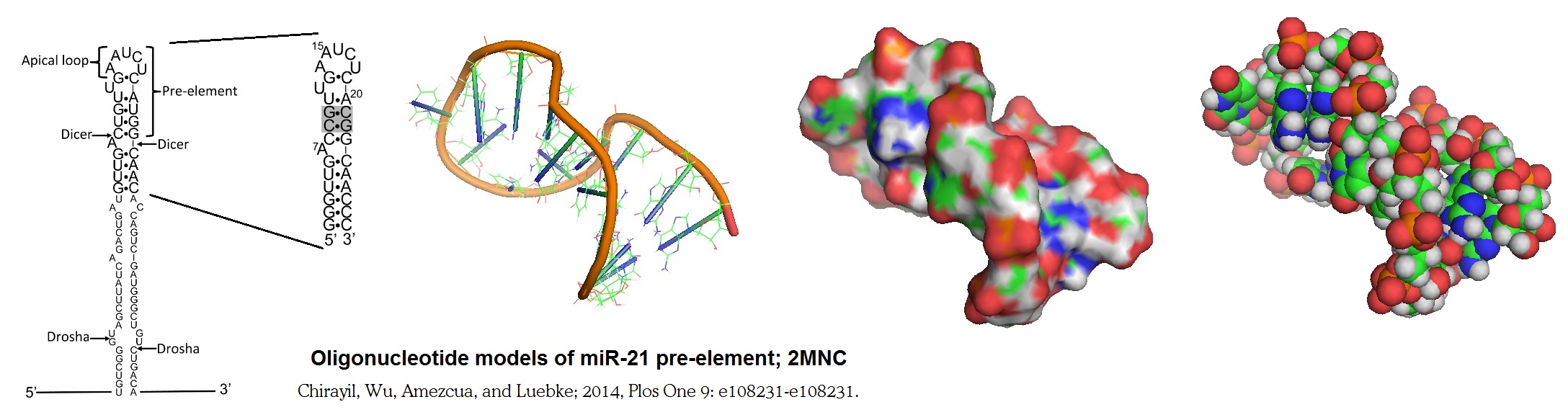
Figure 2: Oligonucleotide model of miR-21 pre-element. Chirayil et al. in 2014 reported the NMR model of an oligonucleotide stem-loop structure based on the pre-element of an oncogenic miRNA, miR-21. Only part of the molecule is shown in the structural model since the secondary structure of the native sequence could only be poorly defined by NMR. The replacement of two putative G•U base pairs with G•C base pairs allowed the researchers to observe the imino proton resonance in the molecule and to solve the structure of the stem-loop. The predicted stem-loop structure is cleaved from the precursor of miR-21 (pre-miR-21) by the nuclease Dicer. This structure is also recognized by the protein complex that converts the primary transcript (pri-miR-21) into the pre-miRNA.
Selected references to review:
miRNAs - http://blog-biosyn.com/2013/08/30/are-micrornas-the-new-players-in-metabolism-and-metabolic-disorders/;
DGCG8 - http://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=ShowDetailView&TermToSearch=54487;
TRBP - http://www.phosphosite.org/proteinAction.do?id=15651&showAllSites=true).
BSI blog: http://blog-biosyn.com/2013/08/30/are-micrornas-the-new-players-in-metabolism-and-metabolic-disorders/
Matthieu P. M. H. Benoit, Lionel Imbert, Andrés Palencia, Julien Pérard, Christine Ebel, Jérôme Boisbouvier and Michael J. Plevin; The RNA-binding region of human TRBP interacts with microRNA precursors through two independent domains. Nucl. Acids Res. (2013), 1-12.
Bredy TW, Lin Q, Wei W, Baker-Andresen D, Mattick JS.; MicroRNA regulation of neural plasticity and memory. Neurobiol Learn Mem. 2011 Jul;96(1):89-94. doi: 10.1016/j.nlm.2011.04.004. Epub 2011 Apr 18.
Sara Chirayil, Qiong Wu, Carlos Amezcua, Kevin J. Luebke; NMR Characterization of an Oligonucleotide Model of the MiR-21 Pre-Element. Plos One 2014, 9, 9, e108231, page 1 to 11. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0108231
Gurtan AM, Sharp PA; The role of miRNAs in regulating gene expression networks. J Mol Biol. 2013 Oct 9;425(19):3582-600. doi: 10.1016/j.jmb.2013.03.007. Epub 2013 Mar 13.
Amy E. Pasquinelli; MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nature Reviews Genetics 13, 271-282 (April 2012) | doi:10.1038/nrg3162. http://www.ncbi.nlm.nih.gov/pubmed/22411466
Wilson RC, Tambe A, Kidwell MA, Noland CL, Schneider CP, Doudna JA; Dicer-trbp complex formation ensures accurate mammalian microrna biogenesis. . Mol Cell. (2015) 57 p.397.
---...---