Digital PCR
Digital PCR (dPCR) enables accurate, highly sensitive quantification of nucleic acids (DNA/RNA). Traditional PCR is an end-point analysis that is semi-quantitative because the amplified product is detected by agarose gel electrophoresis. Real-time PCR (or qPCR) uses fluorescence-based detection to allow the measurement of accumulated amplified product as the reaction progresses. qPCR requires normalization to controls (either to a reference or to a standard curve), allowing only relative quantification.

Furthermore, variations in amplification efficiency may affect qPCR results. Digital PCR builds on traditional PCR amplification and fluorescent-probe–based detection methods to provide highly sensitive absolute quantification of nucleic acids without the need for standard curves. In the Droplet Digital™ PCR System, a PCR sample is partitioned into 20,000 droplets. After amplification, droplets containing target sequence are detected by fluorescence and scored as positive, and droplets without fluorescence are scored as negative. Poisson statistical analysis of the numbers of positive and negative droplets yields absolute quantitation of the target sequence.
There are following describes some of the most relevant areas of application such as.
- Copy Number Variation (CNV)
- Rare Sequence Detection
- Gene Expression
- Single-Cell Analysis
- Pathogen Detection and Microbiome Analysis
- Next-Generation Sequencing (NGS)
Bridged Nucleic Acids (BNA) Technology
Natural nucleic acids have a higher degree of liberty in their chemical structure. This characteristic is thermodynamically unfavorable for DNA-DNA and RNA-RNA double strand formation (hybridization) and is often subject to degradation by both endonucleases and exonucleases. Improving binding affinity (hybridizing capability) is yet unresolved for highly sensitive gene-targeting applications.
Bridged nucleic acid 2',4'-BNANC (2'-O,4'-aminoethylene bridged nucleic acid) is a compound containing a six-member bridged structure with an N-O linkage. This novel nucleic acid analogue can be synthesized and incorporated into oligonucleotides. When compared to the earlier generation of LNA, BNA was found to possess:
- Higher binding affinity against an RNA complement
- Excellent single-mismatch discriminating power
- Enhanced RNA selective binding
- Stronger and more sequence selective triplex-forming characters
- Stronger nuclease resistance to endo and exo-nucleases, even higher than the S(p)-phosphorothioate analogue.
BNA for applications in antisense and antigene technologies.
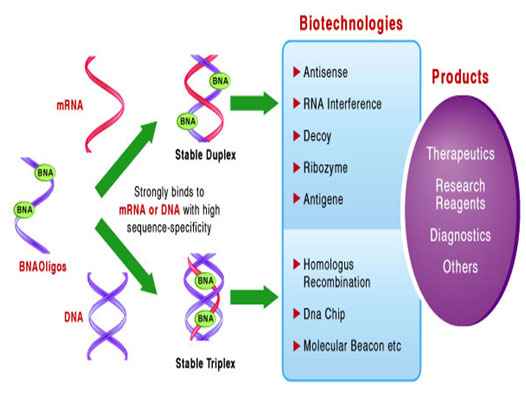
BNA can be easily incorporated into oligonucleotide strands. This feature allows for designing BNA hybrid oligos to satisfy the need for very high and sequence-specific hybridization with nucleic acids. Additionally, BNA possesses a strong nuclease-resistant property. Due to these outstanding properties, Bio-Synthesis Inc. now offers the third generation BNA (BNANC).
BNA Oligonucleotides Synthesis:
BNA oligos allow greater flexibility in the design of primers and probes ideal for the detection of short RNA and DNA targets. . They can be mixed with DNA, RNA and other nucleic acid analogs using standard phosphoramidite synthesis chemistry. BNA oligonucleotides are also easily labeled or modified with standard oligonucleotide tags such as DIG, fluorescent dyes, biotin, amino-linkers etc.
Multi Functional Bridged Nucleic Acid (BNA), An Alternative aspect to LNA and PNA
A novel bead-based suspension assay using Bridged Nucleic Acid, BNA-NC, probes allows quantitative detection of DNMT3A p.R882C/H/R/S mutations. The comparison with LNA based probes revealed the superior hybridization characteristics of the BNA based probes. The researchers demonstrated for the first time the benefit of BNA-NC probes coupled to fluorescently labeled beads for quantitative detection of DNMT3A R882 mutations. This type of assay adds to the list of molecular diagnostics techniques used for the analysis of biological markers in genomic and proteomic research. The specific diagnosis and monitoring of diseases enables detection and evaluation of disease risks for individual patients. By analyzing the specifics of a patient’s disease, molecular diagnostics offers the promise to enable and optimize personalized medicine.
Probe Design for BNA/DNA Probes.
- Primers are designed to allow for the amplification of a DNA sequence fragment that contains the mutated sequence codon. One of the primers is labeled with biotin.
- First, human genomic DNA is extracted from blood.
- The exon 23 of the human DNMT3A gene is amplified using the selected forward and reverse primer.
- To determine the exact sequence the purified and amplified DNA can be sequenced using Big Dye terminator cycle-sequencing.
- Next, the exon 23 DNMT3A fragments are amplified from either genomic or plasmic DNA samples using a 5’-biotinylated forward primer.
- Genotyping is performed with the BNA-NC modified oligonucleotide probes connected to microsphere beads, specific for the wild type or the mutant alleles, by direct hybridization.
- The captured DNA fragment containing biotin on its 5’-end is detected with the help of streptavidin-phycoerythrine (SAPE) in the hybridization buffer using the LabScan200 flow platform from Luminex (USA).
Advantages of the BNA Technology
- Improved hybridization selectivity and specificity over PNA & LNA
- Ideal for the detection of short RNA and DNA targets
- Facilitate Tm normalization
- Increased thermal duplex stability of duplexes
- Increased thermal stability of triplexes
- Superior antisense inhibition and potency
- Strand invasion enable detection of "hard to access" sample
- Flexible probe designs regardless of GC content
- Easily adaptable to many DNA or RNA detection system
- Capable of single nucleotide discrimination
- Resistance to exonuclease and endonucleases resulting in high biological stability for in vivo and in vitro application
Applications of BNA in modern research
- Antigene inhibition
- Gapmer antisense research
- In vivo, in vitro delivery
- Biosensor and more..
- Bio-Synthesis, home of BNATM oligonucleotides
- Inhibition of RNA function
- Real-time/ qPCR
- SNP detection /allele specific PCR
- RNAi research
- In situ hybridization
- DNAzymes and Ribozymes
References
- Boettger LM et al. (2012). Structural haplotypes and recent evolution of the human 17q21.31 region. Nat Genet 44, 881–885. PMID: 22751096
- Karlin-Neumann G et al. (2011). Probing copy number variations using Bio-Rad's QX100™ Droplet Digital™ PCR System. Bio-Rad Bulletin 6277.
- Soo Jin Kim, Hongbo Zhao, Swanand Hardikar, Anup Kumar Singh, Margaret A. Goodell, and Taiping Chen; A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. December 12, 2013; Blood: 122 (25).
- Shivarov V, Ivanova M, Naumova E; Rapid Detection of DNMT3A R882 Mutations in Hematologic Malignancies Using a Novel Bead-Based Suspension Assay with BNA-NC(NC) Probes. PLoS One. 2014 Jun 10;9(6):e99769. doi: 10.1371/journal.pone.0099769. eCollection 2014
- 1.Zon, G.; Geiser, T. G., “Phosphorothioate oligonucleotides: chemistry, purification, analysis, scale-up and future directions”, Anticancer Drug Des. 1991, 6, 539-568.
- 2.Prakash, T. P.; Bhat, B., “2'-Modified oligonucleotides for antisense therapeutics”, Curr. Top. Med. Chem. 2007, 7, 641-649.
- 3.Manoharan, M., “2'-carbohydrate modifications in antisense oligonucleotide therapy: importance of conformation, configuration and conjugation”, Biochim. Biophys. Acta 1999, 1489, 117-130.
- 4.Dellinger, D. J.; Sheehan, D. M.; Christensen, N. K.; Lindberg, J. G.; and. Caruthers, M. H., “Solid-phase chemical synthesis of phosphonoacetate and thiophosphonoacetate oligodeoxynucleotides”J. Am. Chem. Soc., 2003, 125, 940950.
- 5.Summerton, J., Weller, D., “Morpholino antisense oligomers: design, preparation, and properties” Antisense Nucleic Acid Drug Dev., 1997, 7:187-195.
- 6.Hyrup, B.; Nielsen, P. E., “Peptide nucleic acids (PNA): synthesis, properties and potential applications”, Bioorg. Med. Chem. 1996, 4, 5-23.
- 7.Stein, C.A; Cohen, J. S., “Oligodeoxynucleotides as inhibitors of gene expression: a review”, Cancer Res., 1988, 48, 2659-2668.
- 8.Sanghvi, Y. S., “A status update of modified oligonucleotides for chemotherapeutics applications”, Curr. Protoc. Nucleic Acid Chem., 2011, Chapter 4, Unit 4.1.1-22.
- 9.Prakash, T. P.,“An overview of sugar-modified oligonucleotides for antisense therapeutics”, Chem. Bioliers., 2011, 8, 1616-1641.
- 10.Yamamoto, T.; Nakatani, M.; Narukawa, K.; Obika, S., “Antisense drug discovery and development”, Future Med. Chem. 2011, 3, 339-365.
- 11.Veedu, R. N.; Wengel, J., “Locked nucleic acids: promising nucleic acid analogs for therapeutic applications”, Chem. Bioliers., 2010, 7, 536-542.
- 12.Rahman, S.M.; Seki, S.; Utsuki, K.; Obika, S.; Miyashita, K.; Imanishi, T., “2',4'-BNA(NC): a novel bridged nucleic acid analogue with excellent hybridizing and nuclease resistance profiles”, Nucleosides Nucleotides Nucleic Acids., 2007, 26, 1625-1628.
- 13.Rahman, S. M.; Seki, S.; Obika, S.; Yoshikawa, H.; Miyashita, K.; Imanishi, T., “Design, synthesis, and properties of 2',4'-BNA(NC): a bridged nucleic acid analogue”, J. Am. Chem. Soc., 2008, 130, 4886-4896.
- Yamamoto, T.; Harada-Shiba, M.; Nakatani, M.; Wada, S.; Yasuhara, H.; Narukawa, K.; Sasaki, K.; Shibata, M.-A.; Torigoe, H.; Yamaoka, T.; Imanishi, T.; Obika, S., “Cholesterol-lowering Action of BNA-based Antisense Oligonucleotides Targeting PCSK9 in Atherogenic Diet-induced Hypercholesterolemic Mice”, Mol. Ther. Nucleic Acids, 2012, 1, e22.
- 1.Miyashita, K.; Abdur Rahman, S. M.; Seki, S.; Obika, S.; Imanishi, T., "N-Methyl substituted 2',4'-BNANC: a highly nuclease-resistant nucleic acid analogue with high-affnity RNA selective hybridization
- 2.Yamamoto, T.; Harada-Shiba, M.; Nakatani, M.; Wada, S.; Yasuhara, H.; Narukawa, K.; Sasaki, K.; Shibata, M.-A.; Torigoe, H.; Yamaoka, T.; Imanishi, T.; Obika, S., “Cholesterol-lowering Action of BNA-based Antisense Oligonucleotides Targeting PCSK9 in Atherogenic Diet-induced Hypercholesterolemic Mice”, Mol. Ther. Nucleic Acids, 2012, 1, e22.
- 3.Abdur Rahman, S. M.; Sato, H.; Tsuda, N.; Haitani, S.; Narukawa, K.; Imanishi, T.; Obika, S., " RNA interference with 2',4'-bridged nucleic acid analogues", Bioorganic & Mdicinal Chemistry 18 (010) 3474-3480
- Tsuyoshi Yamamoto, et al.: Superior Silencing by 2',4'-BNA NC-based Short Antisense Oligonucleotides Compared to 2',4'-BNA/LNA-based Apolipoprotein B Antisense Inhibitors; Journal of Nucleic Acid, Vol. 2012
- T. Yamamoto et al.: Cholesterol-lowering Action of BNA-based Antisense Oligonucleotides Targeting PCSK9 in Atherogenic Diet-induced Hypercholersterolemic Mic; Molecular TherapyNucleic Acid Researech. 2012.
- M. Nishida. et al.: Synthesis, RNA selective hybridization and high nuclease resistance of an oligonucleotide containing novel bridged nucleic acid with cyclic urea structure; ChemComm 2012.
- S.M. Abdur Rahman et al.: Highly Stable Pyrimidine-Motif Triplex Formation at Physiological pH Values by a Bridged Nculeci acid Analogues; Angeu. Chem. Int. Ed. 2007.
- S.M. Abdur Rahman et al.: Design, Synthesis, and Properties of 2',4'-BNANC: A Bridged Nculeic acid Analogue; JACS, 2008.