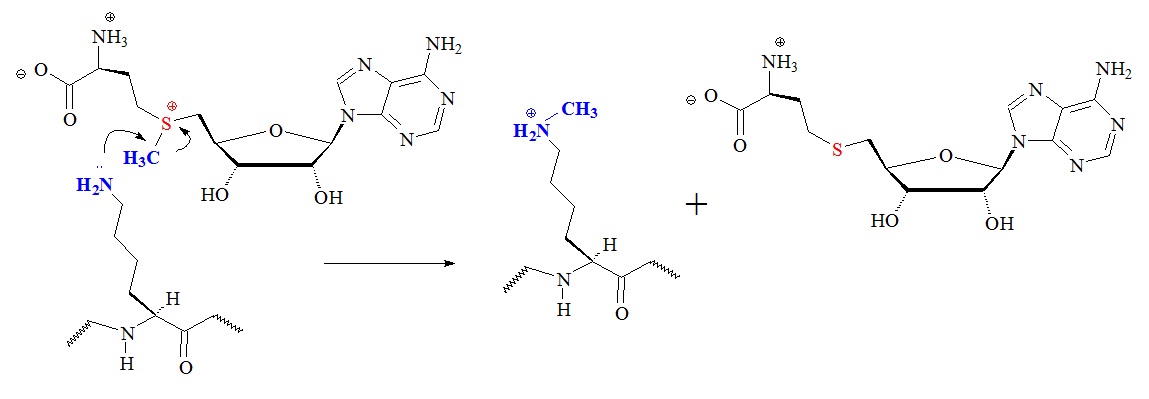
Figure 2: Proposed general chemical mechanism of lysine modifications catalyzed by S-adenosyl-L-methionine (AdoMet)-dependent histone lysine methyltransferases. Apparently SET domain, Dot/KMT4, and protein arginine methyltransferases use a similar mechanism for the transfer of methyl groups from the donor AdoMet.
S-adenosyl-L-methionine {SAM or AdoMet; S-(5’-deoxy-5’-adenosyl)-L-methionine)} is a condensation product of adenosine and L-methionine involving the replacement of the phosphate group (–OPO3H2) of adenylic acid by –S+(CH3)-CH2CH2CH(NH3+)CO2 of methionine. The replacing group is a sulfonium compound bearing a methyl group that is transferred in transmethylation reactions, sometimes also called an active methionine.
The SET domain was first recognized as a conserved sequence in three Drosophila melanogaster proteins. These proteins were a modifier of position-effect variegation, called Suppressor of variegation 3-9 or Su(var)3-9, the Polycomb-group chromatin regulator Enhancer of zeste (E(z)), and the trithorax-group chromatin regulator trithorax (Trx). The domain is approximately 130 amino acids long and was characterized in 1998.
Many SET-domain proteins have now been found in all eukaryotic organisms studied. The histone H3 lysine 79 methyltransferase DOT1L/KMT4 has been found to promote oncogenic pattern of gene expression through binding with several MLL fusion partners found in acute leukemia but its normal function in mammalian gene regulation is poorly understood.
Protein arginine methyltransferases (PRMTs) are encoded in mammalian genomes and catalyze three types of arginine methylation; monomethylation and two types of dimethylation. Methylation of arginine residues in proteins is an abundant modification assumed to be important for transduction, gene transcription, DNA repair and mRNA splicing, among many others.
Transduction refers to any process by which a cell can convert a signal from itself to another cell, or the process by which DNA is transferred from one bacterium to another by a virus. Three types of methylarginine species have been found to exist:
ω‑NG‑monomethylarginine (MMA),
ω‑NG,NG‑asymmetric dimethylarginine (ADMA) and
ω‑NG,N’G‑symmetric dimethylarginine (SDMA).
Furthermore, recent studies have linked these modifications to carcinogenesis and metastasis and studies using newer sequencing technologies have not generally found alterations to the PRTMs. PRTMs are ubiquitously expressed and are involved in important cellular processes that affect cell growth, proliferation and differentiation. The deregulation of these enzymes is thought to cause the pathogenesis of several diseases, possibly including cancer.
The demethylation mechanism for the monoamine oxidase demethylase family LSD1/KDM1 is illustrated in figure 3.
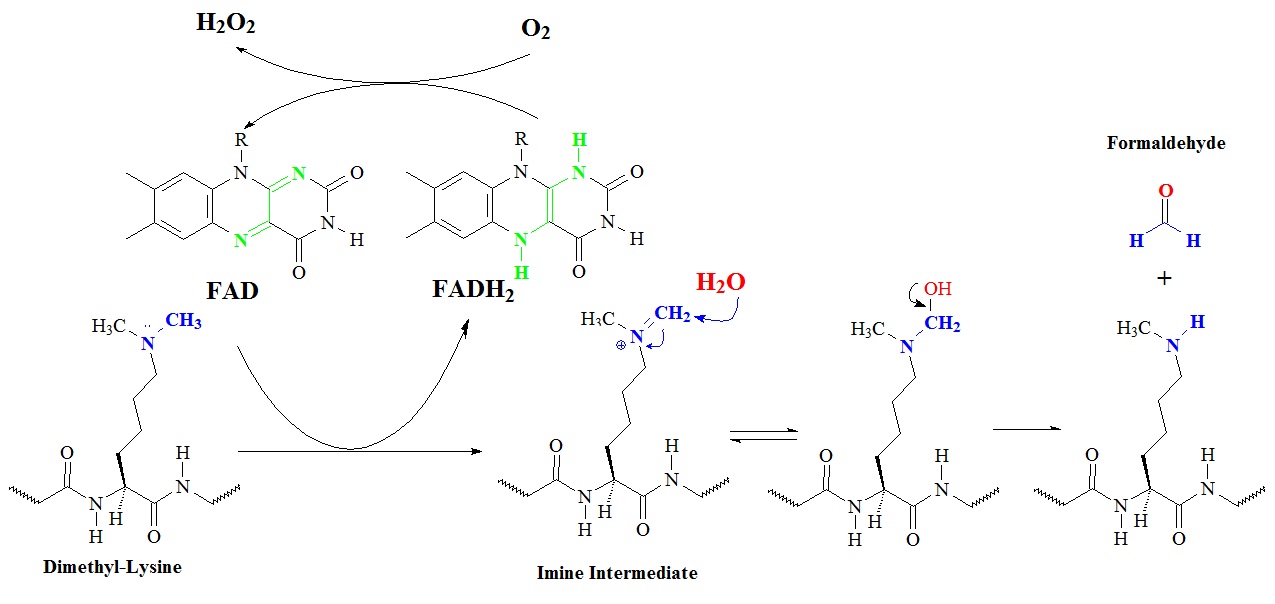
Figure 3: Proposed general chemical mechanism of demethylase enzyme LSD1/KDM1. FAD dependent demethylation, for example of Lys-4 of histone H3, proceeds through the hydrolysis of an iminium ion following a two oxidation of the amine by the flavin moiety. R = ribosyl adenine dinucleotide.
The enzyme lysine (K)-specific demethylase 1A or LSD1 is encoded by the KDM1A gene and is a member of the monoamine oxidase family. This enzyme can demethylate mono- and di-methylated lysines. LSD1 can demethylate histone 3, lysines 4 and 9 (H3K4 and H3K9). However, the precise mechanism is still controversial. Structural studies of LSD1 in complex with histone analog suicide inhibitors allowed Culhane and Cole in 2007 to elucidated part of the catalytic mechanism. Histone 3 peptides were used to create modified peptide analogs called suicide inhibitors that contained propargylamine, aziridine, and cyclopropylamine groups on the epsilon position of lysine 4.
Yang et al in 2007 reported the crystal structure of human LSD1 with a propargylamine-derivatized H3 peptide covalently tethered to FAD, depicted in figure 4. The analysis of the structure showed that H3 adopts three consecutive gamma-turns, enabling an ideal side chain spacing that places its N terminus into an anionic pocket and positions methyl-Lys4 near FAD for catalysis. Furthermore the scientists could show that the LSD1 active site cannot productively accommodate more than three residues on the N-terminal side of the methyllysine. This fact explained the enzymes H3-K4 specificity. This structure of LSD1-bound H3 may act as a model allowing the design of potent LSD1 inhibitors with therapeutic potential.
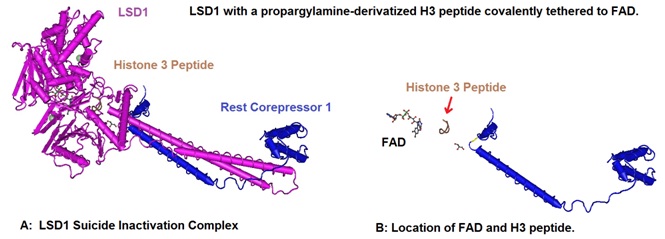
Figure 4: Structural basis of histone demethylation by LSD1 revealed by suicide inactivation. Yang et al in 2007 solved the crystal structure of human LSD1 with a propargylamine-derivatized H3 peptide covalently tethered to FAD (A). H3 adopts a protein fold of three consecutive gamma-turns. This protein domain enables an ideal side chain spacing that places its N terminus into an anionic pocket and positions methyl-Lys 4 near FAD for catalysis. The location of FAD and the modified H3 peptide is illustrated in B. The LSD1 protein structure was deselected to allow a closer look at the FAD and H3 peptide location. The structural data for 2UXN was downloaded from the NCBI structure database and rendered using Cn3D 4.3 to create the models.
Iron (II) dependent a-ketoglutarate oxigenases can hydroxylate alkylgroups directly through a hydroxyl radical. This reaction is thought to provide a plausible path to demethylation of quaternary ammonium salts as indicated in figure 5.
.jpg)
Figure 5: Proposed catalytic mechanism for the iron (II) dependent demethylation of trimethyl-lysine substrates. The reaction proceeds through an iron (II), α-ketoglutarate, O2 derived hydroxyl radical oxidation of the methyl C-H bond.
The enzymes called jumonji histone demethylases or JHDMs are conserved in organisms ranging from yeast to humans and are capable of demethylating trimethyl-lysine residues. JHDMs belong to the Fe (II)/oxoglutarate-dependent dioxygenase or hydroxylase superfamily. The proposed mechanism by which these enzymes catalyze the demethylation of trimethylated lysine residues with the help of a quaternary complex containing 2-oxoglutarate, Fe (II), and the conserved residues His-188, Glu-190, and His-276 is illustrated in figure 6. The JMJD2A/KDM4A numbering according to Allis et al. 2007 is used.
.jpg)
Figure 6: Proposed catalytic mechanism for JHDM enzymes for the demethylation of tri-methylated lysine residues. In the active site of the enzyme, the conserved residues His-188, Glu-190, and His-276 chelate the Fe (II). The first chemical step is an electron transfer from Fe (II) to molecular oxygen generating a superoxide radical and Fe (III). The hypothetical model assumes that the hydrophobic active site assists to generate high oxidation states. Nucleophilic attack of the activated oxygen at the ketone carbon of 2-oxoglutarate results in an Fe (IV) peroxyhemiketal bicyclic intermediate. Subsequent decarboxylation results in a succinate, carbon dioxide (CO2), and an iron (IV)-oxo intermediate. The intermediate oxidizes the methyl carbon of the methylated lysine producing a hemiaminal intermediate and regereating iron (II). The resulting hemiaminal is thought to spontaneously decompose to demethylated lysine and formaldehyde. However, unlike the LSD1/KDM1 mechanism, this mechanism does not require a lone electron pair on the ε-nitrogen of the methylated lysine substrate.
In recent years the importance of chromatin modifications in many aspects of cell biology has become apparent and several enzyme families that modify histones have now been identified. The majority of these proteins are conserved throughout evolution. To minimize the confusion in naming these enzymes Allies et al. in 2007 proposed a new nomenclature for all the characterized members of the families of lysine demethylases, acetyltransferases, and lysine methyltransferases.
The enzymes have been given more generic names that reflect
(1) the type of enzymatic activity they perform and
(2) the type of residue they modify.
Table 1 contains a list of lysine demethylases now called KDMs (K-demethylases).
Table 1: K-Demethylases (KDMs; Formerly Lysine Demethylases)(Allis et al. 2007).
Histone acetyl-transferases (HATs or KATs) catalyze the transfer of the acetyl moiety from acetyl-coenzyme A (acetyl-CoA or AcCoA) to the ε-amino group of histone lysine residues. The result of this reaction is an acetylated lysine side chain and a free CoA. AcCoA is the sole acetylgroup donor for protein acetylation and its concentration in cells affects the level of protein acetylation. CoA is a coenzyme containing pantothenic acid, adenosine 3’-phosphate 5’-pyrophosphate, and cysteamine and is a key intermediate in the mitochondrial metabolism of pyruvate.
HATs are grouped into three main groups or families
(1) the glucosamine-6-phosphate N-acetyltransferase or
Gcn5-related N-acetyltransferase (GNAT) family
(founding member in yeast Gcn5/ScKAT2), and
(2) the monocytic leukemia 3, also known as MYST3
(MYST; founding members MOZ, Ybf2/Sas2, and Tip60) family,
and
(3) the transcriptional coactivators p300 and the cyclic adenosine
mono-phosphate response element-binding protein (CREB)
histone acetyltransferase (p300/CBP) family.
The structure of acetyl-coenzyme A is illustrate in figure 7 and the proposed mechanism for class I/II/IV histone acetyltransferases catalyzing the transfer of the methyl group from the acetylgroup donor AcCoA to the lysine residue is depicted in figure 8.
.jpg)
Figure 7: Acetyl-coenzyme A (AcCoA), an important intermediate in the metabolism of pyruvate, fatty acids and many amino acids. Pyruvate generated in the cytosol during glycolysis is transported across the mitochondrial memebranes to the matrix wher it reacts with HSCoA to form carbon dioxide and acetyl CoA. The reaction is catalyzed by the enzyme pyruvate dehydrogenase which is a soluble component of the matrix. The mitochondria contain an outer membrane, an inner membrane with extensive cristae, the inter-membrane space, and the matrix with small granules, as was revealed by electron microscopy. The matrix contains the mitochondrial DNA and ribosomes.
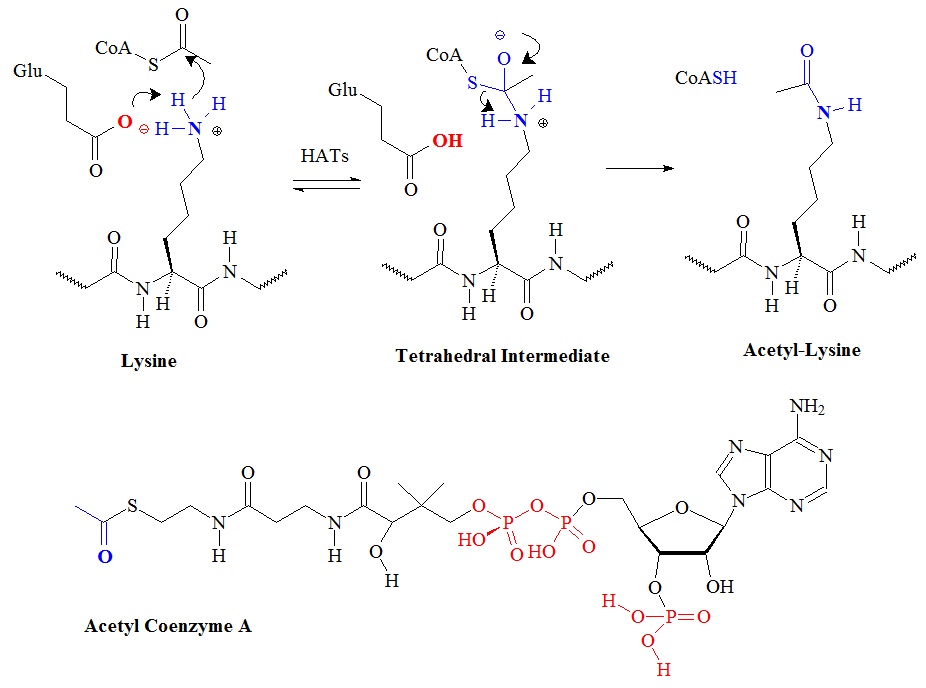 Figure 8: Proposed chemical mechanism of histone acetyl-transferases.
Figure 8: Proposed chemical mechanism of histone acetyl-transferases.
Histone deacetylases remove the acetyl group form lysine side chains in histone proteins. Class I/II/IV histone deacetylases (HDACs) have homologous active-sites and are thought to proceed through a catalytic mechanism in which the two active-site His-Asp dyads work as a general acid-base catalytic pair. The proposed chemical mechanism is illustrated in figure 9.
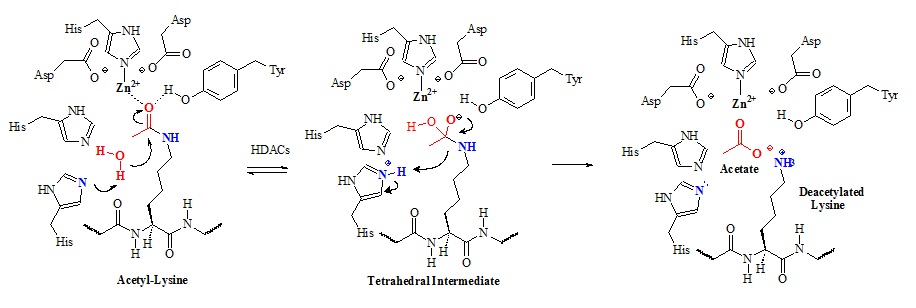
Figure 9: Proposed chemical mechanism of class I/II/IV histone deacetylases (HDACs). In this mechanism an active-site metal ion and Tyr-306 (HDAC8 numbering) polarize the carbonyl by coordinating to the acetyl oxygen. The metal appears to be zinc (II) but could be iron (II) as well. The metal ion is coordinated by Asp-178, His-180, and Asp-267. Structural studies suggested that the metal ion also coordinates and activates the water molecule for attack of the acetyl carbonyl carbon. Apparently, both His-142 and His-143 activates the water molecule for nucleophilic attack of the acetyl carbonyl carbon. Subsequently, the tetrahedral intermediate collapses to form acetate and the lysine products.
Table 1 contains a list of lysine demethylases now called KDMs (K-demethylases).
Table 1: K-Demethylases (KDMs; Formerly Lysine Demethylases)(Allis et al. 2007).
Histone acetyl-transferases (HATs or KATs) catalyze the transfer of the acetyl moiety from acetyl-coenzyme A (acetyl-CoA or AcCoA) to the ε-amino group of histone lysine residues. The result of this reaction is an acetylated lysine side chain and a free CoA. AcCoA is the sole acetylgroup donor for protein acetylation and its concentration in cells affects the level of protein acetylation. CoA is a coenzyme containing pantothenic acid, adenosine 3’-phosphate 5’-pyrophosphate, and cysteamine and is a key intermediate in the mitochondrial metabolism of pyruvate.
HATs are grouped into three main groups or families
(1) the glucosamine-6-phosphate N-acetyltransferase or Gcn5-related
N-acetyltransferase (GNAT) family
(founding member in yeast Gcn5/ScKAT2), and
(2) the monocytic leukemia 3, also known as MYST3
(MYST; founding members MOZ, Ybf2/Sas2, and Tip60)
family, and
(3) the transcriptional coactivators p300 and the cyclic adenosine
mono-phosphate response element-binding protein (CREB)
histone acetyltransferase (p300/CBP) family.
Table 2 contains a list of lysine acetyl-transferases now called KATs (K-acetyl-transferases).
Table 2 : K-Acetyl transferases (KATs; Formerly Lysine Acetyl-transferases) (Allis et al. 2007).
Table 3 contains a list of lysine methyl-transferases now called KMTs (K-methyl-transferases).
Table 3 : K-methyl transferases (KMTs; Formerly Lysine Methyl-transferases) (Allis et al. 2007).
Reference
Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y.; New nomenclature for chromatin-modifying enzymes. Cell. 2007 Nov 16;131(4):633-6.
Jeffrey C. Culhane and Philip A. Cole; LSD1 and The Chemistry of Histone Demethylation. Curr Opin Chem Biol. Oct 2007; 11(5): 561–568. Published online Sep 11, 2007. doi: 10.1016/j.cbpa.2007.07.014, PMCID: PMC2112775. NIHMSID: NIHMS33479.
Darnel, J., Lodish, H., Baltimore D.; Molecular Cell Biology. 2nd edition. Scientific American Books. W.H. Freeman and Company, New York, 1990.
Shane C Dillon, Xing Zhang, Raymond C Trievel, and Xiaodong Cheng; The SET-domain protein superfamily: protein lysine methyltransferases Genome Biol. 2005; 6(8): 227.
Fred Dyda, David C Klein, and Alison B Hickman; GCN5-Related N-Acetyltransferases: A Structural Overview. http://www.ncbi.nlm.nih.gov/books/NBK2231/
Rice JC, Allis CD; Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001 June; 13(3):263-73.
Sabrina Schroeder, Tobias Pendl, Andreas Zimmermann, Tobias Eisenberg, Didac Carmona-Gutierrez, Christoph Ruckenstuhl, Guillermo Mariño, Federico Pietrocola, Alexandra Harger, Christoph Magnes, Frank Sinner, Thomas R Pieber, Jörn Dengjel, Stephan J Sigrist, Guido Kroemer, Frank Madeo; Acetyl-coenzyme A: A metabolic master regulator of autophagy and longevity. Autophagy 10:7, 1335–1337; July 2014; © 2014 Landes Bioscience.
Brian C. Smith and John M. Denu; Chemical mechanisms of histone lysine and arginine modifications. Biochim Biophys Acta. Jan 2009; 1789(1): 45–57.
David J. Steger, Martina I. Lefterova, Lei Ying, Aaron J. Stonestrom, Michael Schupp, David Zhuo, Adam L. Vakoc, Ja-Eun Kim, Junjie Chen, Mitchell A. Lazar, Gerd A. Blobel, and Christopher R. Vakoc; DOT1L/KMT4 Recruitment and H3K79 Methylation Are Ubiquitously Coupled with Gene Transcription in Mammalian Cells. Mol Cell Biol. Apr 2008; 28(8): 2825–2839.
Lawrence M. Szewczuk, Jeffrey C. Culhane, Maojun Yang, Ananya Majumdar, Hongtao Yu, and Philip A. Cole; Mechanistic Analysis of a Suicide Inactivator of Histone Demethylase LSD1. Biochemistry. Jun 12, 2007; 46(23): 6892–6902.
Yang M, Culhane JC, Szewczuk LM, Gocke CB, Brautigam CA, Tomchick DR, Machius M, Cole PA, Yu H; Structural Basis of Histone Demethylation by Lsd1 Revealed by Suicide Inactivation. Nat.Struct.Mol.Biol. (2007) 14 p.535.
Yanzhong Yang & Mark T. Bedford; Protein arginine methyltransferases and cancer. Nature Reviews Cancer 13, 37-50 (January 2013).