Real time detection of influenza virus strain H7N9
The polymerase chain reaction (PCR) allows for the detection of viral DNA or RNA. Whereas the real-time polymerase reaction (RT-PCR) enables the quantitative detection of a virus such as the influenza virus. End-point quantitative RT-PCR can be used to detect the presence of the influenza virus strain H7N9 in infected individuals.
The development of the polymer chain reaction (PCR), for which Kary B. Mullis received a Nobel Prize in 1993 together with Michael Smith, has completely revolutionized the detection of RNA and DNA. PCR based methods now allow the detection of the amplified product at the end-point of the reaction or while the reaction is occurring. The real-time polymerase reaction (RT-PCR), sometimes also called the quantitative chain reaction (qPCR), is a technique based on the polymerase chain reaction (PCR), which can be used to amplify and simultaneously quantify specific DNA molecules.
PCR enables both, detection and quantification of one or more specific DNA sequences in a sample. Detected quantities are reported either as absolute copy numbers or as relative amounts when normalized to DNA input or additional normalizing genes. Real-Time chemistries allow the detection of the PCR amplification products during the early phases of the reaction. The measurement of the reaction kinetics in the early phases of the PCR reaction provides a distinct advantage over traditional PCR detection methods. Usually, traditional methods use agarose gels for detection of PCR amplification products at the final phase or end-point of the PCR reaction. RT-qPCR or RT-PCR can be used to detect viral DNA or RNA.
Classically respiratory viruses have been identified using viral cultures in a variety of permissive cell lines. However, viral cultures are hard to maintain because of the need to rapidly inoculate clinical samples into multiple cell lines to achieve optimal sensitivity. In addition, many rapid immune-enzymatic assays have been developed by many companies to allow detection of influenza virus A and B antigens and HRSV antigens. Unfortunately, the accuracy of these tests depends on many variables, and their sensitivity has generally been lower than that of viral cultures. Therefore PCR assays have now been developed for many respiratory viruses, allowing their detection of small amounts of viral nucleic acid.
Influenza (Flu) viruses occur in two main types: Types A and B. The influenza A and B viruses that routinely spread in people, the human influenza viruses, are responsible for seasonal flu epidemics each year.
Influenza A viruses can be classified into sub-types depending on the genes that code for the surface proteins of the virus strain. However, during a flu season, different types and subtypes of influenza can circulate and cause illness.
According to the CDC human infections with a new avian influenza A (H7N9) virus were first reported in China in March 2013. Most of these infections are believed to result from exposure to infected poultry or contaminated environments, since H7N9 viruses have also been found in poultry in China. Most patients infected with human H7N9 have had severe respiratory illness, with about one-third resulting in death. The first case outside of China was in Malaysia and was reported on February 12, 2014. The case was detected in a traveler from an H7N9-affected area of China. According to the CDC the new H7N9 virus strain has not been detected in people or birds in the United States.
The Influenza A Virus Genome
The influenza virus belongs to the Orthomyxoviridae family composed of six different RNA viruses. This virus is responsible for the common flu in birds, mammals, and humans. The estimated death toll in the United States on average is 50,000 people annually. Symptoms usually include fever, headache, and nasal discharge which can develop into more obstructive pulmonary and heart problems including cardiac failure and bacterial pneumonia. The Influenza A virus genome is contained on eight single (non-paired) RNA strands that code for eleven proteins (HA, NA, NP, M1, M2, NS1, NEP, PA, PB1, PB1-F2, PB2). The total genome size is 13,588 bases. The segmented nature of the genome allows for the exchange of entire genes between different viral strains during cellular cohabitation. Hemagglutinin (HA) determines the extent of the infection of the host organism. During infection influenza viruses bud from the apical surface of polarized epithelial cells (e.g. bronchial epithelial cells) into the lumen of lungs. The infection primarily occurs in the lungs since HA is cleaved by tryptase clara which is restricted to lungs. Clara cells are dome-shaped cells with short microvilli found in the small airways (bronchioles) of the lungs. However HAs of H5 and H7 pantropic avian virus subtypes can be cleaved by furin and subtilisin-type enzymes, allowing the virus to grow in other organs as well.
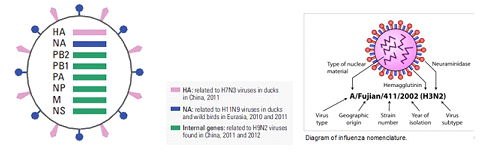
(Source: Science Volume 340 12 April 2013 page 129 – 130 and Wikepidia.)
The eight RNA segments of the viral genome
Real-time PCR (RT-PCR) based assays have been developed in many laboratories now and can be used to detect the different viral strains. The following tables contain lists of real-time detection assays for human H7N9 which has been made possible with the generous contributions of sequence data and viruses by the People's Republic of China, and through the contribution of protocols and validation data from OFFLU network laboratories.
Detection of RNA segment H7 [Hemagglutinin]
Detection of RNA segment N9 [neuraminidase]
Detection of RNA segment M [matrix proteins]
$ detection either predicted by none/few mismatches in primer/probes with template OR validated with H7N9 RNA.
* of primers and probe sequences to novel H7N9 sequence.
# other dyes and quenchers are also expected to work. Indicated dye/quencher combinations are used for these protocols in the contributing laboratories.
& Geelong (AAHL) supplied reagents to ASEAN partners.
Assay Protocol 
Real-time RT-PCR Protocol for the Detection of Avian Influenza A(H7N9) Virus 8 April 2013 Updated on 15 April 2013 Updated on 15 April 2013
The WHO Collaborating Center for Reference and Research on Influenza at the Chinese National Influenza Center, Beijing, China, has made available attached real-time RT-PCR protocol for the detection of avian influenza A(H7N9) virus. For further information please contact us at:gisrs-whohq@who.int
It is strongly recommended that all unsubtypeable influenza A specimens should be immediately sent to one of the six WHO Collaborating Centres for Influenza in the Global Influenza Surveillance and Response System (GISRS)1 for testing and analysis
1 http://www.who.int/influenza/gisrs_laboratory/collaborating_centres/list/en/
The protocol was developed by and belongs to the WHO Collaborating Centre in Beijing. It is made available for emergency use as a service to the public health. It is not for commercial development or for profit. .
- Purpose
To specifically detect avian influenza A(H7N9) virus using real-time RT-PCR with specific primers and probes targeting the matrix, H7 and N9 genes.
- Materials and equipments
- Real-time fluorescence quantitative PCR analysis system
- Bench top centrifuge for 1.5mL Eppendorf tubes
- 10, 200, 1000μL pipettors and plugged tips
- Vortex
- QIAGEN RNeasy Mini Kit
- AgPath one-step RT-PCR kit
- The specific primers and probes for the H7and N9 genes are summarized in the table below. In addition, the use of a primer and probe targeted M gene and house-keeping gene such a RNP is recommended for typing all influenza A virus and internal control in the tests.
Table of PCR primers and probes
Note: FluA and RNase primer/probe sets were from published WHO protocol provided by CDC, Atlanta.
- Other materials: RNase-free 1.5mL eppendorf tubes, RNase-free 0.2mL PCR tubes, powder-free disposables latex glove, goggles, headgear, shoe cover, tips for pipettors, β- thioglycol, 70% alcohol.
- Biosafety
The lysis of the specimen (500 μL lysis buffer with 200 μL clinical samples is recommended) should be to be carried out in a BSL-2 facility with BSL-3 level personal protection equipment. Subsequent procedures can be performed in a BSL-2 laboratory which has separate rooms including reagent preparation area, specimen preparation area and amplification/detection area. The DNA-free area is the clean area and the area of amplified DNA is the dirty area. The work flow is from clean to dirty areas.
- Procedures
- Nucleic acid extraction
The procedure is performed in a BSL-2 biohazard hood in the specimen preparation area according the manufacturer. Elution of the RNA using a final volume of 50 μL H2O is recommended.
- Quality control parameters
Negative control: Sterile water is extracted as a negative control at the same as the nuclear acid extraction of the other specimens. Reagent blank control: RNase free H2O.
Positive control: RNA of the A(H7N9) virus provided. Internal positive control: ribonucleoprotein (RNP) is recommended.
- The reaction system preparation
- (1) Thaw the RT-PCR Master Mix, primers and probes at room temperature in the reagent preparation area of the BSL-2 facility.
- (2) Prepare reaction mixture. Different primer pairs and probes should be prepared in the different tubes respectively. For each reaction:
| Components |
volume(μL) |
| 2× RT-PCR Master Mix |
12.5 |
| primer-forward(40μM) |
0.5 |
| primer-reverse(40μM) |
0.5 |
| Probe (20 μM) |
0.5 |
| 25xRT-pcr enzymes mix |
1 |
| Template RNA |
5.0 |
| RNase Free H2O |
5 |
| Total |
25 |
- Aliquot the reaction mixture into 0.2mL PCR tubes or a 96-well PCR plate as 20μL per tube and label clearly.
- Add five μL of the template RNA for the negative control, test specimens, or positive control into the separate tubes with the reaction mixture in a BSL-2 biohazard hood in the specimen preparation area.
- Load the tubes in the PCR cycler for Real-time RT-PCR detection and use the following programme for cycling:
- 45°C 10min
- 95°C 10min
- 95°C 15s
- 60°C 45s
Return to the 3th step, and perform 40 cycles
- Result analysis:
The results are determined if the quality controls work.
- The specimen is negative if the value of Ct is undetectable,
- The specimen is positive if Ct value is ≤38.0.
- It is suggested that specimens with a Ct higher than 38 are repeated. The specimen can be considered positive if the repeat results are the same as before i.e. Ct is higher than 38. If the repeat Ct is undetectable, the specimen is considered negative.
- Criteria for quality control:
- The result of the negative control should be negative.
- The Ct value of positive control should not be more than 28.0.
- Otherwise, the test is invalid.
- Troubleshooting
- False positives may be due to environmental contamination if there is amplification detected in the negative control and reagent blank control. The unidirectional work flow must be strictly obeyed. The following measures should be taken should there be false positives: ventilate the labs, wash and clean the workbench, autoclave centrifuge tubes and tips, and use fresh reagents.
- RNA degradation should be taken into consideration if the Ct value of the positive control is more than 30. All materials should be RNase-free.
- Cautions
- In order to avoiding nucleic acid cross-contamination, add the negative control to the reaction mixture first, then the specimen, followed by the positive control respectively.
- Dedicated equipment for each area including lab coats, pipettors, plugged tips and powder-free disposal latex glove are required.
- Follow the instructions for maintenance of the incubator, PCR cycler and pipettors. Calibration should be performed every 6 months.
- Protocol Use Limitations
These protocols were optimized using the quantitative one-step probe RT-PCR (AgPath one-step RT-PCR kit ) that have been shown to produce comparable results on 96-well format thermocycler systems such as Stratagene QPCR instruments (MX3000®or MX3005®).
Real time detection of influenza virus H7N9