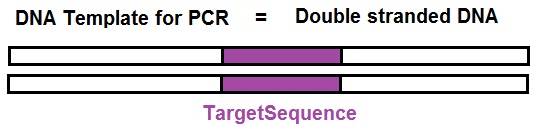
What is needed to amplify a segment of DNA?
1. “Template”: First, we need a few molecules of DNA that include the DNA segment or sequence we want to amplify. This target sequence is called the “template”. Just a trace amount of this DNA fragment is needed.
2. “Primers”: Next, we need PCR primers, two of them. PCR primers are short pieces of single-stranded DNA (ssDNA) that match the sequences at either end of the target DNA segment or sequence. Primers are needed to enable the start of the DNA synthesis. A primer is a short segment of DNA that can bind to a longer sequence template strand and allows the DNA synthesis to get started.


3. “Polymerase”: Furthermore, an enzyme that catalyzes the production of DNA copies is needed as well. Since modern PCR involves high temperature steps, a heat-resistant DNA polymerase is used. In most contemporary PCR, Taq polymerase from Thermus aquaticus is used.
4. “Nucleotides”: Also, a supply of nucleotides is also needed. The polymerase uses the nucleotides to make new DNA. Four deoxynucleoside triphosphates dATP, dCTP, dGTP, and dTTP together with magnesium (Mg2+), salts and pH buffer are added.
5. “Thermocycler”: To allow PCR to occur a PCR machine or a temperature cycling instrument is needed as well. Since the PCR process requires the cycling of several temperatures, the PCR instrument is also called a thermocycler. Without a thermocycler reaction tubes containing the PCR reaction mixture have to be moved between three incubators at different temperatures every few minutes.

For PCR to occur, the DNA strands need to be separated. Heating the template to 90 °C for a minute or two will allow for this to happen.
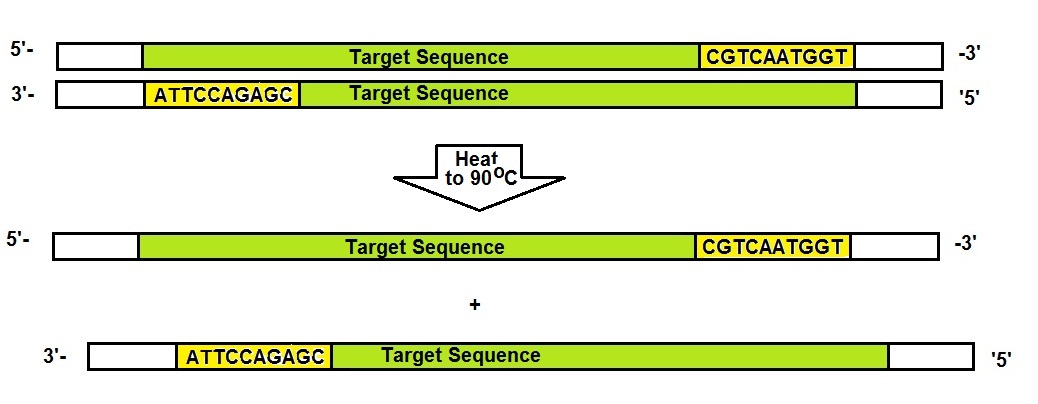

Next, to allow the primers present in the PCR mixture to bind or anneal to the complementary target sequence the temperature needs to be lowered to approximately 50 to 60 °C. In general, PCR primers are 15 to 20 bases long. A shorter primer is used here for illustration.


In the next step, the temperature is maintained at approximately 70 °C for one or two minutes. At this temperature, the polymerase can elongate new DNA strands. The elongation starts at the primers from the 5’ to the 3’ end for both strands. Finally, two partial double-stranded pieces of DNA are synthesized. The two new stands are not as long as the original template and both are missing a piece at the end where the synthesis started.
.jpg)

Next, the cycle of temperature changes is repeated resulting in four (4) partial double-stranded DNA pieces.
As the temperature cycles or cycling continues through the PCR, the single-stranded overhangs are ignored and are outnumbered by DNA segments containing only the target sequence.

The temperature cycling is repeated over and over again. The result is that the major product is the double-stranded target sequence with flush ends.
Each cycle doubles the amount of DNA. In theory, if the amplification is 100 % there should be a doubling of the PCR product in every cycle. The next figure illustrates the exponential increase in DNA products.
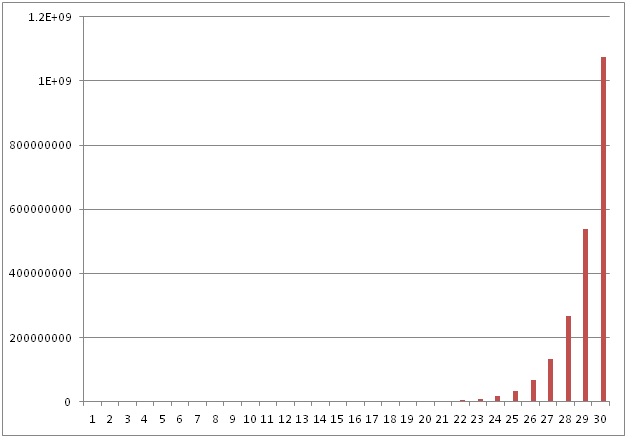

However, in a real PCR experiment the efficiency of target amplification is usually less than 100%.
The equation that describes the exponential amplification of PCR is:
Xn = Xo * (1 + Ex)n
where
Xn = number of target molecules at cycle n
Xo = initial number of target molecules
Ex = effifiency of target amplification
n = number of cycles.
In real-time PCR fluorescence monitoring of DNA, amplification allows to determine DNA concentrations. Absolute quantification can be achieved with the construction of standard curves using known amounts of DNA.
References
http://www.biosyn.com/newsletters/real-time-pcr-probes.html
http://www.biosyn.com/tew/The-Polymerase-Chain-Reaction-or-PCR.aspx
http://www.brandeis.edu/wanghlab/papers/naturemethods.pdf
BustinS. ; Absolute Quantification Of mRNA Using Real-Time Reverse Transcription Polymerase Chain Reaction Assays. Journal Of Molecular Endocrinology, Vol.25, P.169-193, 2000.
Kuchel and Ralston; Biochemistry, 2ndediton. Schaum’s outlines. McGRAW –HILL.
Mullis K.; In Methods In Enzymology, Vol.155, 335, 1987.
Pattyn F. Rtprimersdb : The Real-Time PCR Primer And Probe Database, Major Update 2006. Nucleic Acids Res 34, D684-8, 2006.
Pattyn F. Rt primers db : The Real-Time PCR Primer And Probe Database, Major Update 2006. Nucleic Acids Res 34, D684-8, 2006.
Schmittgen Td. Real-Time PCR Quantification Of Precursor And MatireMicrorna. Methods 44, 31-8, 2008.
Scipioni A. A Sybr Green Rt-Pcr Assay In Single Tube To Detect Human And Bovinenoroviruses and control for inhibition. Virol J. 5(1):94, 2008.
http://en.wikipedia.org/wiki/Reverse_transcription_polymerase_chain_reaction