2'-O, 4'-C-Ethylene-bridged Nucleic Acids (ENA) are promising modified nucleic acids in which ethylene is bridged at the furanose sugar ring at the 2'-O and 4'-C ends to form a less strained six-membered ring with C3'-endo conformation. ENA gapmers effectively recruited RNase H, and ENA act as splice modulators such as exon-skipping antisense oligonucleotides and antigene oligonucleotides. Also, clinical trials are currently being conducted using ENA oligonucleotide to treat Duchenne muscular dystrophy (DMD). The aim of the treatment is to skip the normal exon 45 of the dystrophin gene to prevent the occurrence of a stop codon in exon 45. This will restore the functional dystrophin. This technology is owned by Daiichi Sankyo, and they are conducting a Phase II clinical study in Japan.[1]
Background: Koizumi et al. developed the building block for bicyclo 2′-O, 4′-C-ethylene-bridged nucleic acid (ENA) and compared the properties of 2’,4’-BNA/LNA, a third-generation nucleic acid. Also, further development on synthetic methods to improve the productivity of the monomer has been reported.[2] Interestingly, apart from high thermodynamic stability, ENA showed enhanced nuclease resistance while retaining the binding affinity strength of other 2’,4’-BNA/LNA. ENA restricts the single-strand flexibility of RNA by forcing the North conformation of sugar puckering within nucleosides due to the 2′-O,4′-C-ethylene-linkage like 2’,4’-BNA/LNA. Studies have shown that 2′,4′-bridged nucleic acids can improve the ability of complementary strands to bind to duplexes. However, a recent report by J. Kawakami et al discovered that not all 2′,4′-bridged nucleic acids can lead to duplex stabilization. When comparing the properties of 2’,4’-C-bridged 2’-deoxynucleotide (CRN) with ENA, it was found that the introduction of one CRN into DNA/RNA duplexes destabilized them. [3,4]
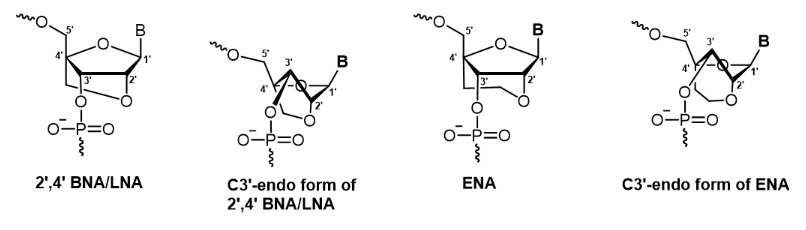
Figure1: Structure of 2’,4’-BNA/LNA, ENA and their C3’- endo form
What makes ENA a better choice?
1. Binding affinity: ENA exhibits higher binding affinity to complementary RNA and can match that of 2′,4′-BNA/LNA (ΔTm=+3∼5 °C per modification).[5]
2. Nuclease resistance: ENAs exhibited greater nuclease resistance than natural DNA and 2′,4′-BNA/LNA. Including one ENA in oligonucleotides significantly enhances their resistance to exonucleases, surpassing that of 2′,4′-BNA/LNA.[6]
3. Thermal stability: ENA exhibits high thermal stability. A single modification of 2’O, 4’-C-ethyleneguanosines resulted in a 10-fold increase in the binding constant of the DNA/RNA duplex.[7]
4. Chimeric RNA and ENA show greater efficacy compared to phosphorothioate oligodeoxynucleotides in causing exon 19 skipping in dystrophin mRNA.[8]
5. ENA exhibits a great ability to form a triplex structure with double-stranded DNA.[6]
These results indicate that ENA is more suitable as an antisense oligonucleotide and is expected to have better antisense activity than 2′,4′-BNA/LNA. These remarkable characteristics distinguish it from other nucleic acids and render it an attractive choice for developing antisense therapies. Considering its exceptional chemical stability, ENA holds significant potential for developing oligonucleotides that are resistant to nuclease degradation, thereby enhancing their efficacy and promoting their clinical utility. The unique properties of ENA make it a compelling area of research for those involved in the field of antisense therapeutics.
References
1. M. Matsuo et al., Genes, 2017, 8, 67.
2. M. Michida et al., Organic Process Research and Development, 2022, 26, 1289-1307.
3. J. Kawakami et al., Nucleosides, Nucleotides & Nucleic acids, 2024, 1, 57-64.
4. M. Koizumi, Biological and Pharmaceutical Bulletin, 2004, 27, 453-456.
5. M. Koizumi et al., Bioorganic & Medicinal Chemistry Letters, 2002, 12, 73–76.
6. M. Koizumi et al., Bioorganic & Medicinal Chemistry, 2003, 11, 2211–2226.
7. M. Koizumi et al., Nucleic Acids Symposium Series, 2005, 49, 171-172.
8. M. Matsuo et al., Oligonucleotides, 2004, 14, 33-40.