L-DNA is a mirror-image form of the naturally occurring d-conformation of DNA. L-DNA forms a left-helical double-helix but has similar physical characteristics in regards to solubility, duplex stability, and selectivity as D-DNA. However, L-DNA cannot form stable duplexes with d-DNA.
L-DNA is useful for the design of functional nucleic acid probes, for example in biological applications. Advancements made in solid-phase and liquid-phase synthesis of DNA now allows incorporation of L-DNA into regular DNA as well as into nucleic acid-based conjugates or other nucleic acid-based constructs.
In 1980, Arnott et al. observed a ladder-like DNA (L-DNA) structure in the X-ray diffraction pattern obtained from calf thymus DNA saturated with intercalating [(bipy)2Pt(en)]2+. The analysis of the x-ray pattern revealed that when saturation with the intercalator, the DNA double helix is unwound into a linear, duplex arrangement similar to a ladder. Therefore this DNA was named L-DNA. The intercalator molecule is inserted in the structure into every second, alternate site. In each Watson-Crick base-pair of L-DNA, one base is in syn, but the partner in anti orientation. The syn base is associated with the C3’-endo sugar pucker, and the anti with a c2’-endo sugar pucker.
In 2003, Cherrak et al. reported the structure of the complex of d(C1m8G2C3G4C5LG6LC7G8C9G10)2 containing L-nucleotides and 8-methylguanine. Solving the structure of the complex revealed that the nucleotides act cooperatively to promote a left-handed helix under physiological salt conditions.
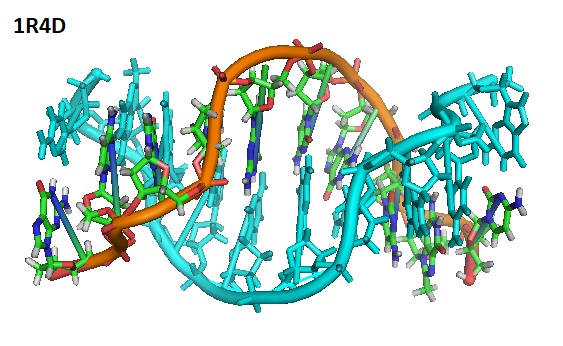
Structure of the d(C1m8G2C3G4C5LG6LC7G8C9G10)2 containing L-nucleotides and 8-methylguanine.
In 2006, Hauser et al. reported that L-DNA allows analyzing different marker types on a single universal microarray platform.
And in 2007, Kim et al. showed that incorporating L-DNA into the stem region of a molecular beacon reduces intra- and intermolecular stem invasions. Furthermore, this incorporation increased the melting temperature, and the selectivity to its target is also improved, leading to enhanced bio-stability.
More recently, in 2011, Corradini et al. reviewed how chiral, or mirror image based molecules, influence DNA binding, and interactions, as well as the major structural factors underlying the formation of stable heterochiral complexes. Complexes studied were obtained by incorporation of modified nucleotides into natural duplexes, or by hybridization between homochiral opposite strands.
In summary, the incorporation of modified DNA such as L-DNA or BNAs into regular DNA allows the generation of a large variety of DNA helices at low salt concentrations by manipulating internal factors such as sugar configuration, duplex length, nucleotide composition, and base methylation.
Reference
Cherrak I, Mauffret O, Santamaria F, Hocquet A, Ghomi M, Rayner B, Fermandjian S; L-nucleotides and 8-methylguanine of d(C1m8G2C3G4C5LG6LC7G8C9G10)2 act cooperatively to promote a left-handed helix under physiological salt conditions. Nucleic Acids Res. (2003) 31 p.6986-95.
Corradini R, Sforza S, Tedeschi T, Marchelli R.; Chirality as a tool in nucleic acid recognition: principles and relevance in biotechnology and in medicinal chemistry. Chirality. 2007 May 5;19(4):269-94.
D'Alonzo D, Guaragna A, Palumbo G.; Exploring the role of chirality in nucleic acid recognition. Chem Biodivers. 2011 Mar;8(3):373-413. doi: 10.1002/cbdv.201000303.
Hauser NC, Martinez R, Jacob A, Rupp S, Hoheisel JD, Matysiak S. Utilising the left-helical conformation of L-DNA for analysing different marker types on a single universal microarray platform. Nucleic Acids Res. 2006 Oct;34(18):5101-11. doi: 10.1093/nar/gkl671. Epub 2006 Sep 20. PubMed PMID: 16990248; PubMed Central PMCID: PMC1636439. [Pubmed]
Kim Y, Yang CJ, Tan W. Superior structure stability and selectivity of hairpin nucleic acid probes with an L-DNA stem. Nucleic Acids Res. 2007 Dec;35(21):7279-87. doi: 10.1093/nar/gkm771. Epub 2007 Oct 24. PubMed PMID: 17959649; PubMed Central PMCID: PMC2175343.
Sanger, W.; 1984. Principles of Nucleic Acid Structure. Springer-Verlag. New York, Berlin, Heidelberg, Tokyo.
---...---