|
|
|
Bio-Synthesis Newsletter - July 2019
|
Tumors are dependent on membrane lipid remodeling
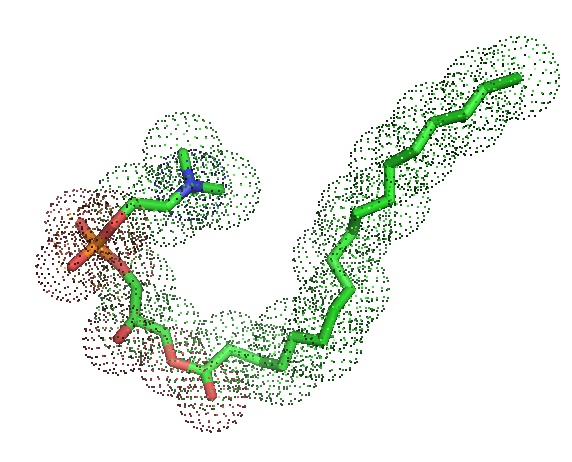 Lysophosphatidylcholine acyltransferase (LPCAT1), when dysregulated in cell tissue links common genetic alterations found in tumors with changes in their metabolism to drive aggressive tumor growth. Bi et al. recently showed that LPCAT1 links specific genetic alterations in cancer with aberrant metabolism and plasma membrane remodeling to drive tumor growth. Cancers driven by growth factor receptor depend on LPCAT1 to shape plasma membrane composition through enhanced saturated phosphatidylcholine content. LPCAT1activates membrane lipid remodeling due to persistent EGFR signaling and regulates phospholipid saturation and oncogenic growth factor signaling. Furthermore, LPCAT1 is frequently upregulated in cancer and is associated with poor patient survival. According to Bi et al., LPCAT1 is a new drug target in a Lysophosphatidylcholine acyltransferase (LPCAT1), when dysregulated in cell tissue links common genetic alterations found in tumors with changes in their metabolism to drive aggressive tumor growth. Bi et al. recently showed that LPCAT1 links specific genetic alterations in cancer with aberrant metabolism and plasma membrane remodeling to drive tumor growth. Cancers driven by growth factor receptor depend on LPCAT1 to shape plasma membrane composition through enhanced saturated phosphatidylcholine content. LPCAT1activates membrane lipid remodeling due to persistent EGFR signaling and regulates phospholipid saturation and oncogenic growth factor signaling. Furthermore, LPCAT1 is frequently upregulated in cancer and is associated with poor patient survival. According to Bi et al., LPCAT1 is a new drug target in a
|
|
Synthetic long RNAs containing terminal 5′‐phosphate groups
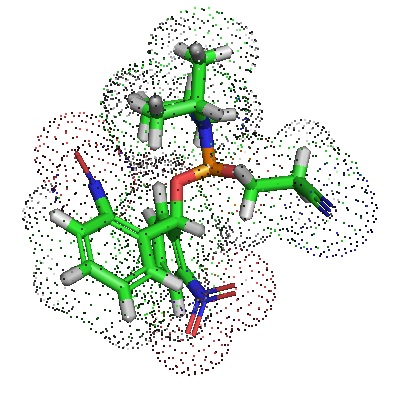 Pradere et al. in 2017 described an easy‐to‐prepare phosphoramidite reagent allowing the chemical introduction of 5′‐terminal phosphate groups into long RNAs. Long or Ubermer RNAs are useful tools in biochemistry and biology usually prepared enzymatically or using the solid‐phase synthesis of multiple RNAs loaded with 5′‐terminal phosphate groups. Template ligation reactions enable concatenation of the modified RNAs. In the reported phosphoramidite, adinitrobenzhydryl group protects the phosphate and serves as an essential lipophilic group for the separation of oligonucleotide by‐products. A brief UV irradiation unmasks the phosphate group quantitatively. The new phosphoramidite allows the preparation libraries of long structured RNAs site‐specifically modified with functional groups. Pradere et al. in 2017 described an easy‐to‐prepare phosphoramidite reagent allowing the chemical introduction of 5′‐terminal phosphate groups into long RNAs. Long or Ubermer RNAs are useful tools in biochemistry and biology usually prepared enzymatically or using the solid‐phase synthesis of multiple RNAs loaded with 5′‐terminal phosphate groups. Template ligation reactions enable concatenation of the modified RNAs. In the reported phosphoramidite, adinitrobenzhydryl group protects the phosphate and serves as an essential lipophilic group for the separation of oligonucleotide by‐products. A brief UV irradiation unmasks the phosphate group quantitatively. The new phosphoramidite allows the preparation libraries of long structured RNAs site‐specifically modified with functional groups.
|
|
Kissing DNA
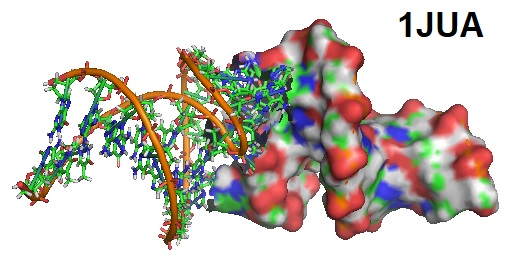 Barbault et al. in 2020 reported the NMR solution structure of a kissing complex formed with two DNA strands. The research group synthesized the deoxyoligoribonucleotide d(CTTGCTGAAGCGCGCACGGCAAG) (dSL1) corresponding to the reverse transcripted sequence of the dimerization initiation site SL1 of HIV- 1(Lai) RNA using phosphoramidite chemistry and found that like its oligoribonucleotide counterpart, dSL1, it dimerized spontaneously in solution allowing solving the solution structure using NMR. Retroviruses have two non-covalently linked copies of genomic RNA near their 5’-end. Immediately upstream from the splice donor site inside the HIV-1 psi-RNA region is a sequence located functioning as the domain for initiation of the genomes dimerization. The kissing complex and the stable dimer are both involved in the HIV-1Lai RNA dimerization process in vitro. Furthermore, the dimerization is activated by the NCp7 protein that converts a transient loop-loop complex into a more stable dimer. The NMR structure revealed a zipper-like motif, A8A9.A16, which positions the surrounding bases more closely to each other, permitting the pairing of the G7 and C17 bases. Barbault et al. in 2020 reported the NMR solution structure of a kissing complex formed with two DNA strands. The research group synthesized the deoxyoligoribonucleotide d(CTTGCTGAAGCGCGCACGGCAAG) (dSL1) corresponding to the reverse transcripted sequence of the dimerization initiation site SL1 of HIV- 1(Lai) RNA using phosphoramidite chemistry and found that like its oligoribonucleotide counterpart, dSL1, it dimerized spontaneously in solution allowing solving the solution structure using NMR. Retroviruses have two non-covalently linked copies of genomic RNA near their 5’-end. Immediately upstream from the splice donor site inside the HIV-1 psi-RNA region is a sequence located functioning as the domain for initiation of the genomes dimerization. The kissing complex and the stable dimer are both involved in the HIV-1Lai RNA dimerization process in vitro. Furthermore, the dimerization is activated by the NCp7 protein that converts a transient loop-loop complex into a more stable dimer. The NMR structure revealed a zipper-like motif, A8A9.A16, which positions the surrounding bases more closely to each other, permitting the pairing of the G7 and C17 bases.
|
|
Capping and decapping enzymes
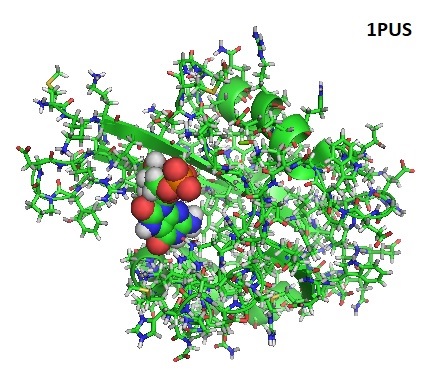 During gene expression capping and decapping of mRNAs occurs. The chemical nature of the 5′ end of RNA determines the stability of RNAs during RNA processing, localization, as well as the translation efficiency. It is possible that this type of modification provides an additional layer of gene regulation called “epitranscriptomic” gene regulation. Enzymes that are known to decap mRNA belong to two different protein families, the Nudixpyrophosphohydrolases and the HIT family of pyrophosphatases. Cellular cofactors cap bacterial as well as eukaryotic transcripts and specific RNA polymerases add cofactors during transcription initiation as well. Mitochondrial RNA binding proteins (RNAP) also cap transcripts with ADP-containing cofactors. Unfortunately, the role of universal RNAP-catalyzed capping is not yet clear, and more studies are needed. During gene expression capping and decapping of mRNAs occurs. The chemical nature of the 5′ end of RNA determines the stability of RNAs during RNA processing, localization, as well as the translation efficiency. It is possible that this type of modification provides an additional layer of gene regulation called “epitranscriptomic” gene regulation. Enzymes that are known to decap mRNA belong to two different protein families, the Nudixpyrophosphohydrolases and the HIT family of pyrophosphatases. Cellular cofactors cap bacterial as well as eukaryotic transcripts and specific RNA polymerases add cofactors during transcription initiation as well. Mitochondrial RNA binding proteins (RNAP) also cap transcripts with ADP-containing cofactors. Unfortunately, the role of universal RNAP-catalyzed capping is not yet clear, and more studies are needed.
|
|
|
|
Bio-Synthesis, Inc.
800 Mario Court, Lewisville, TX 75057, USA
Toll Free: 800.227.0627 | 1.972.420.8505 (Intl.)
|
|
|
|
|
|