The DNA i-motif is a DNA structure in which two parallel-stranded duplexes with C.C(+) pairs are intercalated head-to-tail. Multimeric associations or intra-molecular folding can form the i-motif. However, the formation of the i-motif depends on the number of cytidine tracts, the nucleotide sequences between them, and the biochemical conditions.
The second layer of DNA structures provides regulatory control. B-from DNA stores the genetic information in mammalian cells and humans. A DNA structure called the i-motif formed in cytosine-rich regions of the genome also appears to have regulatory functions.
To probe for the presence of these structures in vivo, Zeraati et al., in 2018, generated and characterized an antibody fragment (iMAB) that recognized the i-motif form with high selectivity and affinity. Utilizing immunofluorescence staining, the research group showed that i-motif structures are present in the nuclei of human cells. The study’s results indicated that i-motif DNA structures form under physiological conditions and are highly cell-cycle specific. The iMab method revealed that the highest level of i-motif formation occurs in the late G1 phase. A high level of transcription and cellular growth characterizes the late G1 stage. Also, the researchers suggested that i-motifs play a regulatory role in the promotors of several proto-oncogenes. Compared to G4 DNA structures, i-motif structures are less stable, transient, and pH-dependent. The reported results indicate that C-rich sequences within the genome can form i-motif structures in the nuclei of human cells and control regulatory functions.
These findings suggest that mutations altering i-motif stability and conformation can affect their regulatory role. However, to confirm this notion, more research is needed.
The i-motif was first observed in 1993 when Gehring et al. solved the DNA oligomer 5'-d(TCCCCC) structure at acid pH using NMR. The structure revealed a four-stranded complex in which two base-paired parallel-stranded duplexes are orientated in an antiparallel fashion so that each base pair is face-to-face with its neighbors (Figure 1). A solution structure of a modified centromeric fragment was solved in 2001 (Figure 2).
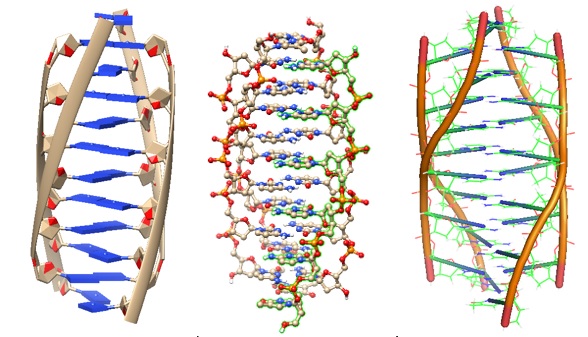
Figure 1: A tetrameric DNA structure with protonated cytosine.cytosine base pairs. PDB ID 225D (Gehring et al. 1993).
Figure 2: Solution structure of a modified human centromeric fragment. PDB ID 1G22 Nonin-Lecomte, S.,Leroy, J.-L. (2001) J Mol Biol 309: 491-506
Reference
Gehring K., Leroy J.L., Gueron M.; A tetrameric DNA-structure with protonated cytosine.cytosine Base-Pairs. Nature. 1993; 363:561–565. [PubMed]
i-Motif Wiki: i-motif DNA - Wikipedia
Nonin-Lecomte S, Leroy JL. Structure of a C-rich strand fragment of the human centromeric satellite III: a pH-dependent intercalation topology. J Mol Biol. 2001 Jun 1;309(2):491-506. doi: 10.1006/jmbi.2001.4679. PMID: 11371167. [Pubmed]
Zeraati, Mahdi; Langley, David B.; Schofield, Peter; Moye, Aaron L.; Rouet, Romain; Hughes, William E.; Bryan, Tracy M.; Dinger, Marcel E.; Christ, Daniel (June 2018). I-motif DNA structures are formed in the nuclei of human cells. Nature Chemistry. 10 (6): 631-637.
---...---
Bio-Synthesis provides a full spectrum of oligonucleotide and peptide synthesis including bio-conjugation services as well as high quality custom oligonucleotide modification services, back-bone modifications, conjugation to fatty acids and lipids, cholesterol, tocopherol, peptides as well as biotinylation by direct solid-phase chemical synthesis or enzyme-assisted approaches to obtain artificially modified oligonucleotides, such as BNA antisense oligonucleotides, mRNAs or siRNAs, containing a natural or modified backbone, as well as base, sugar and internucleotide linkages.
Bio-Synthesis also provides biotinylated mRNA and long circular oligonucleotides.
---...---