Bridged Nucleic Acids = BNAs
Beyond peptide nucleic acids (PNAs) and locked nucleic acids (LNAs)
The near completion of the human genome sequence in 2001 as well as the availability of whole genome sequences for many organisms in recent years, has led to an exponential increase in genomic information in the last decade. The NIH website, http://www.ncbi.nlm.nih.gov/genome, contains a collection of sequences for partial or whole genomes, the number of which is constantly increasing at a high speed. This huge amount of sequence information provides a large source for the design of genetic tools to study molecular details of the genetic information flow in organisms. Scientific evidence shows that humans and other vertebrates are made up by close to or more than 220 different specialized cell types. Each cell type has its own gene regulatory network. To understand an organism at the molecular level the regulation of the genetic information flow will need to be studied. For this purpose new tools have been and are constantly developed. The double helix of DNA is nature’s solution to storing, retrieving, and communicating genetic information of a living organism. Two of many important characteristics of the DNA molecule are the specificity and the reversible nature of the hydrogen bonding between complementary nucleobases. These properties allow the DNA strands of the double helix to unwind and rewind in exactly the same configuration. It is now understood that the cell contains a vast collection of ‘nano-machines’ that control gene regulation. DNA, RNA, proteins, and other molecules make up these nano-assemblies. Control points for gene expression that are studied by scientists include transcriptional regulation, RNA processing, translational control, the stability of mRNA, posttranslational control, and DNA rearrangements among others. These regulatory systems found in prokaryotes and eukaryotes can differ from each other in many details. Once the nature of the DNA and RNA molecules was established, the field of life science realized early on that if specific, single strands of DNA could be synthesized, scientists would be able to study and manipulate the gene sequences of DNA and RNA. This was realized with the development of efficient chemistries including automated instrumentation to allow for the synthesis of DNA and RNA monomers and polymers of increased size including their modifications. Oligonucleotide chemistry has been developed greatly over the past three decades and many advances have been made in the design of DNA, RNA and peptide based molecular tools with increased nuclease resistance.
The double helix exists in multiple conformations which were revealed by X-ray diffraction studies of concentrated DNA solutions that had been drawn out into thin fibers. Two kinds of structures, the B and the A forms of DNA were found. It is thought that the B form corresponds to the average structure of DNA under physiological conditions. Its structure has 10 base pairs per turn, and a wide major groove and a narrow minor groove. The A form on the other hand has 11 base pairs per turn and its major groove is narrower and much deeper than that of the B form. Furthermore, its minor groove is broader and shallower. The B DNA is found in the vast majority of the DNA in the cell. However, A DNA can be found in certain DNA-protein complexes. The non-covalent bonds in double stranded DNA (dsDNA) sequences are formed by Watson-Crick interactions. On the other hand, in triplex forming oligonucleotides the triplex forming strand binds to the duplex via Hoogsteen hydrogen interactions. Structures of the nucleotides and their interaction are shown in the following figure.
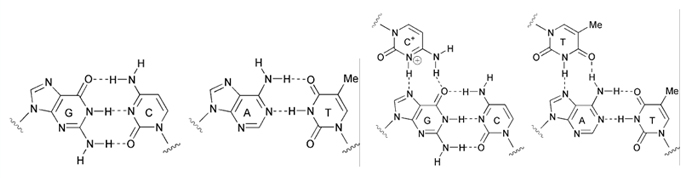 Structures of Watson-Crick-type base pairs, G•C and A•T (left), and Hoogsteen type base triads, C+•G•C and T•A•T (right).
Structures of Watson-Crick-type base pairs, G•C and A•T (left), and Hoogsteen type base triads, C+•G•C and T•A•T (right).
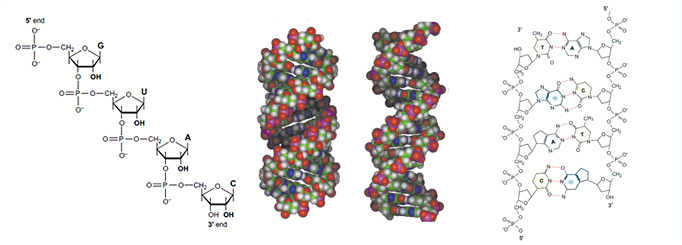
The chemical structures for RNA and DNA are shown below as well as the structures for the A form of RNA and the B-form of DNA.
|
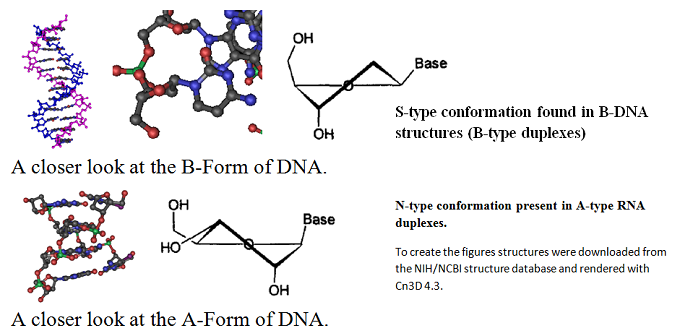
RNA contains ribose rather than 2’-deoxyribose in its backbone. The ribose has a hydroxyl group at the 2’-position. Furthermore, RNA contains the nucleic acid uracil in place of thymine and is usually found as a single polynucleotide chain. While RNA is typically single stranded, RNA chains can frequently fold back on themselves to form base-paired segments between short stretches of complementary sequences. The presence of 2’-hydroxyls in the RNA backbone favors a structure that resembles the A-form structure of DNA. The flexible five-membered furanose ring in nucleotides exists in equilibrium of two conformations of the N- and the S-type.
|
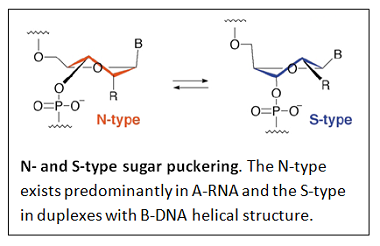
A closer look at the two forms is shown next.
Synthetic oligonucleotides are now important, established tools for life scientists enabling many new applications in molecular biology, genetic diagnostics and are poised to become important tools in the emerging field of molecular medicine as well. While unmodified oligodeoxynucleotides can form DNA:DNA and DNA:RNA duplexes they are sometimes unstable and labile to nucleases. Therefore, a variety of nucleic acid analogs have been developed to enhance high-affinity recognition of DNA and RNA targets, enhancing duplex stability and assisting with cellular uptake. Among the list of these analogs are peptide nucleic acids (PNAs), 2’-fluoro N3-P5-phosphoamidites, 1’, 5’-anhydrohexitol nucleic acids (HNAs) and locked nucleic acids (LNAs) as well as other Bridged Nucleic Acids (BNA). Peptide nucleic acids (PNAs) are synthetic polymers that contain a peptide backbone and nucleic acid bases as side chains within their sequence. These polymers can form strong specific bonds with complementary sequences present in double-stranded DNA (dsDNA). 2’-fluoro N3-P5-phosphoamidites are modified nucleotide analogs that contain fluor in the 2’ position. Hexitol nucleic acids are oligonucleotides built up from natural nucleobases and a phosphorylated 1,5-anhydrohexitol backbone. Anhydrohexitol oligonucleotides can be synthesized using phosphoramidite chemistry and standard protecting groups. LNAs are structurally rigid oligo-nucleotides with increased binding affinities.
A bridged nucleic acid (BNA) is a molecule that can contain a five-membered or six-membered bridged structure. Ideally the bridge is synthetically incorporated at the 2’, 4’-position of the ribose to afford a 2’, 4’-BNA monomer. The monomers can be incorporated into the oligonucleotide polymer structure using standard phosphoamidite chemistry. The goal for the synthesis and use of BNAs is to generate oligonucleotides with (i) equal or higher binding affinity against an RNA complement with excellent single-mismatch discriminating power, (ii) much better RNA selective binding, (iii) stronger and more sequence selective triplex-forming characters, and (iv) with a pronounced higher nuclease resistance, even higher than Sp-phosphorthioate analogues, than regular DNA or RNA oligonucleotides.
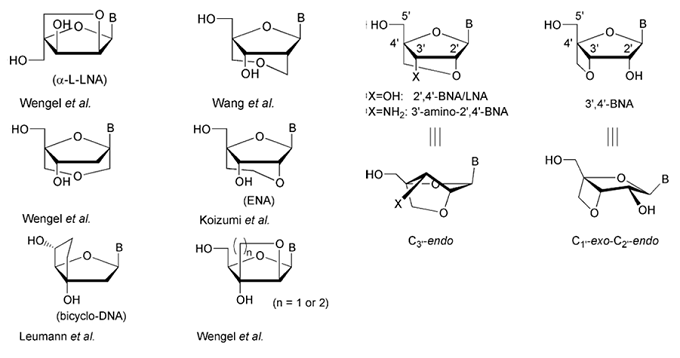
Monomer structures of BNAs. The structures of selected nucleic acid analogs with a bridged sugar moiety are depicted.
While a large number of chemically modified oligonucleotides have been developed during the last few decades, most of these molecules have failed to give the desired response and the search for new molecules with better qualities still continues today. One dramatic improvement was made with the introduction of the bridged nucleic acid, 2’, 4’-BNA (also called LNA). The compound shows a better hybridization affinity for complementary strands, both for RNA and DNA strands, in comparison to unmodified nucleotides. Furthermore the BNA can be used to design sequence selective LNA-oligonucleotide hybrids that are soluble in aqueous solutions and exhibit improved biostability in comparison to natural nucleotides. This BNA monomer has now been widely used in nucleic-acid-based technologies. However, according to Imanishi’s group, there is a need for further development because the nuclease resistance of the LNA is significantly lower than that obtained by phosphorothioate oligonucleotide and the fact that oligonucleotides containing consecutive LNA units or a fully modified oligonucleotide using the analog are very rigid and inflexible. Furthermore, additional research has proven that a kind of LNA-modified antisense oligonucleotide is hepatotoxic.
The scientists Satoshi Obika, Daishu Nanbu, Yoshiyuki Hari, Ken-ichiro Morio, Yasuko In, Toshimasa Ishida, and Takeshi Imanishi in 1997 reported the synthesis of a 2’-O,4’-C-Methyleneneuridine and –cytidine, novel bicylic nucleosides with a fixed C3 - endo sugar puckering. One of these nucleoside analogs is now known as a “locked nucleotide” or LNA. In the ribofuranose, the plane C1’-O4’-C4’ is fixed. The C3’- endo conformation is found in RNA. DNA can adjust and is able to take on both conformations. The exact nomenclature of sugar puckers can be found at
1998
Obika et al. in 1998 (Imanishi’s research group) report the synthesis of bicyclic nucleoside analogues with a fixed N-type conformation, 2'-O,4'-C-methyleneuridine and –cytidine and the incorporated of this analogue into oligonucleotides. The binding efficiency of the modified oligonucleotides to the complementary DNA and RNA as well as the CD spectra of the modified DNA-DNA and modified DNA-RNA duplexes were studied.
Imanishi’s group has synthesized newer generations of BNAs with improved properties, one called 2’4’-BNA-COC, and more recently, one called 2’,4’-BNA-NC. To optimize the length of the bridging moiety the BNA-NC was designed to contain a six-membered bridged structure with an N-O bond in the sugar molecule. The next figures show the chemical structures and 3D ball-and-stick model for the three BNAs.
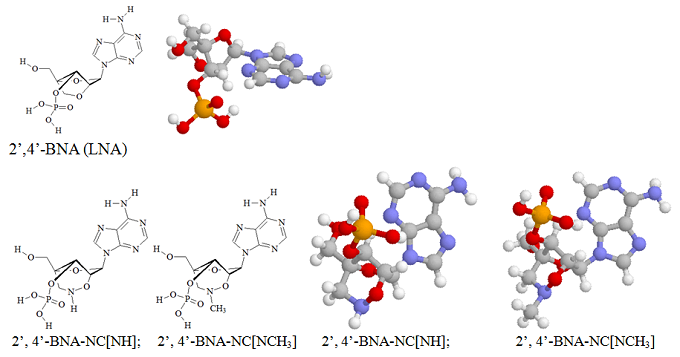
The chemical structures and the energy minimized molecular models for the 2’, 4’-BNA-NC[NH] and 2’, 4’-BNA-NC[NCH3] monomers are illustrated here.
2003
Hari et al. in 2003 developed a novel nucleoside analogue for the effective recognition of CG interruption in a homopurine–homopyrimidine tract of double-stranded DNA (dsDNA). The group succeeded to synthesize a triplex-forming oligonucleotide (TFO) containing a novel 2’,4’-BNA (QB) bearing 1-isoquinolone as a nucleobase. The group used this BNA to investigate the triplex-forming ability and sequence-selectivity of the TFO. Using melting temperature (Tm) measurements, it was found that the TFO-QB formed a stable triplex DNA in a highly sequence-selective manner under near physiological conditions.
2007
Rahman et al. reported in 2007 that oligonucleotides containing 2’, 4’-BNA-NC monomers show an increase in their melting temperature (Tm) of 5.3 to 6.3 °C per modification (ΔT/mod) when investigated by UV melting experiments (Tm measurements).The melting temperature (Tm) is defined as the temperature at which half of the DNA strands are in the double-helical state and half are in the random coil state. The Tm depends on the length of the oligonucleotide molecule and its sequence. The Tm values of duplexes formed by 2’, 4’-BNA-NC[NH] oligonucleotides that contained three or more modifications are higher than those exhibited by the corresponding 2’, 4’-LNA modified oligonucleotides indicating a stronger binding and stability of the duplexes. Both 2’, 4’-BNA-NC[NH] and 2’, 4’-BNA-NC[NCH3] display selective binding to RNA, superior to that of 2’, 4’-LNAs. The researchers used mismatch discrimination studies to evaluate their ability for selective hybridization. The presence of a mismatched base in the target RNA strand resulted in a substantial decrease in the Tm of the duplexes formed. The analysis of formed duplexes by circular dichroism (CD) verified the presence of the A-form by showing the spectra typical for this structure. The A-form of the helical structure is favored because of the conformational restriction of the sugar moiety to the N-form. Furthermore, the BNAs can form triplexes with a higher stability compared to that of LNAs. Applications in antigenic and gene repair technologies require the formation of stable triplexes at physiological pH. Next, to test the BNAs for their resistance to nucleases, oligonucleotides [5’-d(TTTTTTTTTT)-3’] modified with a single 2, 4’-BNA-NC unit were incubated with a 3’-exonuclease (Crotalus adamanteus venom phosphodiesterase, CAVP, Pharmacia) at 37 °C and the amount of intact oligonucleotides remaining were evaluated by RP-HPLC. The results showed that oligonucleotides modified with BNA-NC[NMe] are much more resistant to degradation than LNA modified oligonucleotides.
Rahman et al. in 2007 report on the use of BNA analogues to allow the formation of highly stable pyrimidine-motif triplexes at physiological pH. The formation of a stable triplex DNA molecule at physiological pH values is a highly desirable phenomenon in molecular biology and medicinal chemistry because of its function in the regulation of gene expression, site-specific cleavage of DNA, gene mapping and isolation, the maintenance of folded chromosome conformations, and gene-targeted mutagenesis. The researchers argue that in a pyrimidine-motif triplex DNA, the (homopyrimidine) triplex-forming oligonucleotide (TFO) binds to the homopurine tract of the target duplex DNA. This binding is sequence specific and maintained through Hoogsteen hydrogen bonds to form T·A:T and C+·G:C triads. The formation of the C+·G:C triad depends on the protonation of cytosine, which is only favorable at acidic pH values (pKa=4.5). Therefore, homo-pyrimidine-motif triplexes are extremely unstable at physiological pH values severely limiting their biological application. The group synthesized a novel bridged nucleic acid analogue, 2’,4’-BNANC, and demonstrated that the TFOs composed of 2’,4’-BNANC formed highly stable pyrimidine-motif triplexes at physiological pH values. Furthermore they show that the TFOs eliminate the requirement of placing alternating DNA monomers for optimum efficacy needed when using LNAs. They could show that fully modified TFOs still formed a highly stable triplex. They stated that “these promising properties of 2’,4’-BNANC will be helpful for developing oligonucleotide-based technologies for the postgenome era.”
Also in 2007, Imanishi’s group synthesized oligonucleotides that were modified with a novel BNA analogue, 2’, 4’-BNANC[N–CH3], and compared their properties to oligonucleotides containing 2’,4’-BNA (LNA). They showed that these oligonucleotides have a similarly high RNA affinity but a better selectivity for RNA and a much higher resistance to nuclease degradation. These results suggested that the novel BNA analogue may be particularly useful for antisense approaches when used for the design of antisense oligonucleotides.
2010
Kasahara et al. in 2010 published a paper in where they showed that the capping of the 3’-ends of thrombin binding aptamers (TBAs) with bridged nucleotides increased the nuclease resistances and the stability of the aptamers in human serum. The capping did not affect the binding abilities of the aptamers. The researchers report that the capping could be achieved via a one-step enzymatic process using 2’, 4’-bridged nucleoside 5’-triphosphate and the enzyme terminal deoxynucleotidyl transferase.
In the same year Rahman et al. reported the results of their study in which a number of 2’,4’-BNA- and 2’,4’-BNANC-modified siRNAs were designed and synthesized. The thermal stability, nuclease resistance and gene silencing properties against cultured mammalian cells were evaluated and compared with those of natural siRNAs. The 2’,4’-BNA- and 2’,4’-BNANC-modified siRNAs showed very high Tm values and were remarkably stable in serum samples. Furthermore, the researchers report that these modified oligo-nucleotides showed promising RNAi properties that were equal to those exhibited by natural siRNAs. The thermally stable siBNAs composed of slightly modified sense and antisense strands suppressed gene expression equal to that of natural siRNA. The modifications at the Argonaut (Ago2) cleavage site of the sense strand (9–11th positions from the 5’-end of the sense strand) produced variable results depending on siRNA composition. However, modification at the 10th position diminished siRNA activity. In moderately modified siRNAs, modification at the 11th position displayed usual RNAi activity, while modification at the 9th position showed variable results depending on the composition of the siRNA.
2012
Yamamoto et al. in 2012 demonstrated that BNA-based antisense therapeutics can be used to successfully inhibit hepatic PCSK9 expression which resulted in a strong reduction of the serum LDL-C levels of mice. These findings support the hypothesis that PCSK9 is a potential therapeutic target for hypercholesterolemia. The researchers state that this is the first time they were able to show that BNA-based antisense oligo-nucleotides (AONs) induced a cholesterol-lowering action in hypercholesterolemic mice. Hypercholesterolemia is a metabolic condition in which high levels of cholesterol are present in the blood and where elevated levels of lipids and lipoproteins are observed in the blood as well. If untreated higher levels of total cholesterol will increase the risk for cardiovascular disease in particular coronary heart disease.
We expect that BNAs like LNAs will find a widespread use in antisense oligonucleotide technology, where they can be used to stabilize interactions with target RNA and protect them from the attack by cellular nucleases. Furthermore, they can be used in the field of molecular diagnostics and the newly emerging field of siRNAs. Utilizing these modified nucleotides promises to increase double-stranded RNA stability in serum and decrease off-target effects seen with conventional siRNAs. Next, this oligonucleotide technology has the potential to become a new type of therapy to treat a wide variety of diseases, and BNAs will no doubt play a part in future developments of therapeutic and diagnostic oligonucleotides.
Comparison of various nucleic acid analogs
Rules for the Design of Probes
# You et al. 2006 proposed the following rules to maximize single mismatch discrimination using LNA probes.
Benefits of the BNA technology include:
* Ideal for the detection of short RNA and DNA targets
* Increases the thermal stability of duplexes
* Increases the thermal stability of triplexes
* Capable of single nucleotide discrimination
* Resistant to exo- and endonucleases resulting in high stability in vivo and in vitro applications
* Increased target specificity
* Facilitates Tm normalization
* Strand invasion properties enable the detection of “hard to access” samples
* Compatible with standard enzymatic processes
Applications
- Design and synthesis of RNAaptamers
- siRNA
- antisense probes
- diagnostics
- Isolation
- Microarray analysis
- Northern blotting
- Real-time PCR
- In situ Hybridization
- Functional analysis
- SNP detection
- To use as antigens and many others nucleotide base applications
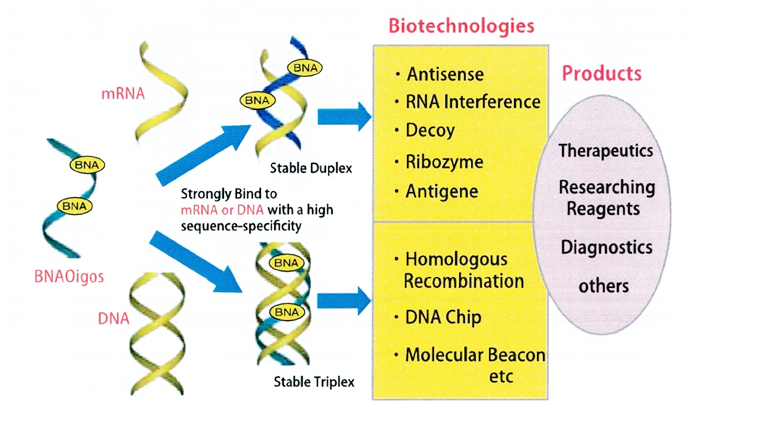
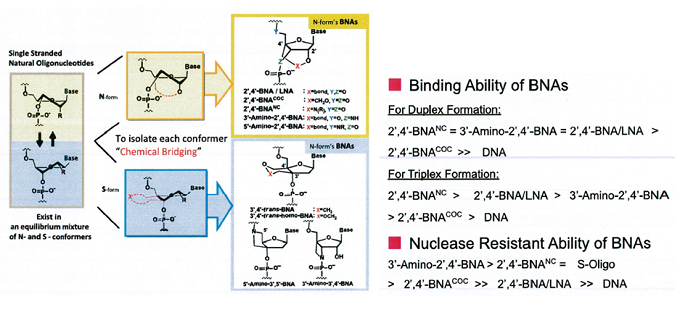
Figures below show graphical illustrations of the function and use of BNAs.
 References
References
Braasch DA, and DR Corey 2001 Locked nucleic acid (LNA): fine-tuning the recognition of DNMA and RNA. Chem Biol 8: 1-7.
Braasch DA, Y Liu, and DR Corey 2002 Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. NAR 30: 5160-5167.
Grunweller A, E Wyszko, B Bieber,R Jahnel, VA Erdmann, and J Kurreck 2003, Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2’-O-methyl RNA, phosphorothioates and small interfering RNA. NAR 31: 3185-3193.
Hendrix C, H Rosemeyer, B De Bouvere, A Van Aerschot, F Seela, and P Herdewijn 1997 1',5'-Anhydrohexitol oligonucleotides: hybridisation and strand displacement with oligoribonucleotides, interaction with RNase H and HIV reverse transcriptase. Eur J Chem 3: 1513–1520.
Yoshiyuki Hari, Satoshi Obika, Mitsuaki Sekiguchi and Takeshi Imanishi; Selective recognition of CG interruption by 20,40-BNA having 1-isoquinolone as a nucleobase in a pyrimidine motif triplex formation. Tetrahedron 59 (2003) 5123–5128.
Hyrup B, and PE Nielson 1996 Peptide nucleic acids (PNA): synthesis, properties and potential applications. Bioorg Med Chem 4:5-23.
Takeshi Imanishi and Satoshi Obika; BNAs: novel nucleic acid analogs with a bridged sugar moiety. CHEM. COMMUN., 2002, 1653–1659.
Johnson MP, LM Haupt, and LR Griffiths 2004 Locked nucleic acids (LNA) single nucleotide polymorphism (SNP) genotype analysis and validation using real-time PCR. NAR 32: e55.
Yuuya Kasahara, Shunsuke Kitadume, Kunihiko Morihiro, Masayasu Kuwahara, Hiroaki Ozaki, Hiroaki Sawai, Takeshi Imanishi, Satoshi Obika; Effect of 3’-end capping of aptamer with various 2’,4’-bridged nucleotides: Enzymatic post-modification toward a practical use of polyclonal aptamers. Bioorganic & Medicinal Chemistry Letters 20 (2010) 1626–1629.
Harleen Kaur, B. Ravindra Babu, and Souvik Maiti; Perspectives on Chemistry and Therapeutic Applications of Locked Nucleic Acid (LNA). Chem. Rev. 2007, 107, 4672-4697.
Tetsuya Kodama,Chieko Matsuo, Hidetsugu Ori, Tetsuya Miyoshi, Satoshi Obika, Kazuyuki Miyashita, Takeshi Imanishi; Design, synthesis, and evaluation of a novel bridged nucleic acid, 2’,5’-BNAON, with S-type sugar conformation fixed by N–O linkage. Tetrahedron 65(2009) 2116–2123.
Koshkin AA, SK Singh, P Nielsen, VK Rajwanshi, R Kumar, M Meldgaard, CE Olsen, and J Wengel 1998 LNA (Locked Nucelic Acid): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 54: 3607-3630.
Kurreck J, E Wyszko, C Gillen, and VA Erdmann 2002 Design of antisense oligonucleotides stabilized by locked nucleic acids. NAR 30: 1911-1918.
Kvaerno L, and J Wengel 1999 Investigation of restricted backbone conformation as an explanation for the exceptional thermal stabilities of duplexes involving LNA (Locked Nucleic Acid): synthesis and evaluation of abasic LNA. Chem Commun 1999, 7: 657-658.
Latorra D, K Campbell, A Wolter, and JM Hurley 2003a Enhanced allele-specific PCR discrimination in SNP genotyping using 3’ locked nucleic acid (LNA) primers. Hum Mut 22: 79-85.
Latorra D, K Arar, and JM Hurley 2003b Design considerations and effects of LNA in PCR primers. Mol Cell Probes 17: 253-9.
McTigue PM, RJ Peterson, and JD Kahn 2004 Sequence-dependent thermodynamic parameters for locked nucleic acid (LNA)-DNA duplex formation. Biochemistry 43: 5388-5405.
Yasunori Mitsuoka, Tetsuya Kodama, Ryo Ohnishi, Yoshiyuki Hari, Takeshi Imanishi and Satoshi Obika; A bridged nucleic acid, 2’,4’-BNACOC: synthesis of fully modified oligonucleotides bearing thymine, 5-methylcytosine, adenine and guanine 2’,4’-BNACOC monomers and RNA-selective nucleic-acid recognition. Nucleic Acids Research, 2009, Vol. 37, No. 4 1225–1238. Note: BNA-COC/DNA and BNA-COC/RNA duplex formation.
Kazuyuki Miyashita, S. M. Abdur Rahman, Sayori Seki, Satoshi Obikaab and Takeshi Imanishi; N-Methyl substituted 2’,4’-BNANC: a highly nuclease-resistant nucleic acid analogue with high-affinity RNA selective hybridization. Chem. Commun., 2007, 3765–3767.
Nielson PE, and G Haaima 1997 Peptide nucleic acid (PNA). A DNA mimic with a pseudopeptide backbone. Chem Soc Rev 26: 73-78.
Nomenclature for polynucleotide chains including for the sugar puckering can be found at:
Satoshi Obika, Daishu Nanbu, Yoshiyuki Hari, Ken.ichiro Morio, Yasuko In, Toshimasa Ishida, and Takeshi Imanishi; Synthesis of 2'-O,4'-C-Methyleneuridine and -cytidine. Novel Bicyclic Nucleosides Having a Fixed C a ,-endo Sugar Puckering. Tetrahedron Letters, Vol. 38, No. 50, pp. 8735-8738, 1997.
Obika S, D Nanbu, Y Hari, J-i Andoh, K-i Morio, T Doi, and T Imanishi 1998 Stability and structural features of the duplexes containing nulcoeside analogs with a fixed N-type conformation. 2’-O, 4’-C methylene ribonucleosides. Tetrahedron Lett 39: 5401-5404.
Satoshi Obika, Mayumi Onoda, Koji Morita, Jun-ichi Andoh, Makoto Koizumi and Takeshi Imanishi; 3’-Amino-2’,4’-BNA: novel bridged nucleic acids having an N3’->P5’ phosphoramidate linkage. Chem. Commun., 2001, 1992–1993. Note: BNA/DNA; BNA/dsDNA.
Satoshi Obika, Yoshiyuki Hari, Mitsuaki Sekiguchi, and Takeshi Imanishi; A 2',4'-Bridged Nucleic Acid Containing 2-Pyridone as a Nucleobase: Efficient Recognition of a C●G Interruption by Triplex Formation with a Pyrimidine Motif. Angew. Chem. Int. Ed. 2001, 40, No. 11, 2079-2081.
Satoshi Obika, Mitsuaki Sekiguchi, Roongjang Somjing, and Takeshi Imanishi; Adjustment of the g Dihedral Angle of an Oligonucleotide P3’!N5’ Phosphoramidate Enhances Its Binding Affinity towards Complementary Strands. Angew. Chem. Int. Ed. 2005, 44, 1944 –1947.
Satoshi Obika, Masaharu Tomizu, Yoshinori Negoro, Ayako Orita, Osamu Nakagawa, and Takeshi Imanishi; Double-Stranded DNA-Templated Oligonucleotide Digestion Triggered by Triplex Formation. ChemBioChem 2007, 8, 1924 – 1928. Note: Triplex triggered cleavage of oligonucleotides.
Satoshi Obika, Hiroyasu Inohara, Yoshiyuki Hari,_ and Takeshi Imanishi; Recognition of T●A interruption by 2’,4’-BNAs bearing heteroaromatic nucleobases through parallel motif triplex formation. Bioorganic & Medicinal Chemistry 16 (2008) 2945–2954.
Satoshi Obika, S. M. Abdur Rahman, Bingbing Song, Mayumi Onoda, Makoto Koizumi, Koji Morita, Takeshi Imanishi; Synthesis and properties of 3’-amino-2’,4’-BNA, a bridged nucleic acid with a N3’->P5’ phosphoramidate linkage. Bioorganic & Medicinal Chemistry 16 (2008) 9230–9237.
Petersen M, and J Wengel 2003 LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol 21:74-81.
Petersen M, CB Nielsen, KE Nielsen, GA Jensen, K Bondensgaard, SJ Singh, VK Rajwanshi, AA Koshkin, BM Dahl, J Wengel, and JP Jacobsen 2000 The conformations of locked nucleic acids (LNA). J Mol Recognition 13: 44-53.
Petersen M, JJ Sorensen, and JT Nielsen 2003 Structural basis for LNA (locked nucleic acid) triplex formation. Presented at the 5th International Congress on Molecular Structural Biology.
S. M. Abdur Rahman, Sayori Seki, Satoshi Obika, Sunao Haitani, Kazuyuki Miyashita, and Takeshi Imanishi; Highly Stable Pyrimidine-Motif Triplex Formation at Physiological pH Values by a Bridged Nucleic Acid Analogue. Angew. Chem. Int. Ed. 2007, 46, 4306 –4309.
S. M. Abdur Rahman, Sayori Seki, Satoshi Obika, Haruhisa Yoshikawa, Kazuyuki Miyashita, and Takeshi Imanishi; Design, Synthesis, and Properties of 2’,4’-BNANC: A Bridged Nucleic Acid Analogue.J. AM. CHEM. SOC. 2008, 130, 4886-4896.
S. M. Abdur Rahman, Hiroyuki Sato, Naoto Tsuda, Sunao Haitani, Keisuke Narukawa, Takeshi Imanishi, Satoshi Obika; RNA interference with 2’,4’-bridged nucleic acid analogues. Bioorganic & Medicinal Chemistry 18 (2010) 3474–3480.
Schulz RG, and SM Gryaznov 1996 Oligo-2'-fluoro-2'-deoxynucleotide N3'-->P5' phosphoramidates: synthesis and properties. NAR 24: 2966-73.
Simeonov A, and TT Nikiforov 2002 Single nucleotide polymorphism genotyping using short, fluorescently labeled locked nucleic acid (LNA) probes and fluorescence polarization detection. NAR 30: e91.
Singh SK, P Nielsen, AA Koshkin, and J Wengel 1998 LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem Commun 4: 455-456.
Tolstrup N, PS Nielsen, JG Kolberg, AM Frankel, H Vissing, and S Kauppinen 2003 OilgoDesign: optimal design of LNA (locked nucleic acid) oligonucleotide capture probes for gene expression profiling. NAR 31: 3758-3762.
Torigoe H, Y Hari, M Sekiguchi, S Obika, and T Imanishi 2001; 2’-O, 4’-C-methylene bridged nucleic acid modification promotes pyrimidine motif triplex DNA formation at physiologic pH. J Biol Chem 276: 2354-2360. Note: TFO for therapeutics.
Hidetaka Torigoe, Osamu Nakagawa, Takeshi Imanishi, Satoshi Obika, Kiyomi Sasaki; Chemical modification of triplex-forming oligonucleotide to promote pyrimidine motif triplex formation at physiological pH. Biochimie 94 (2012) 1032-1040. Note: pyrimidine motif triplex formation by 3’-amino-2’-O,4’-BNA.
Ugozzoli LA, D Latorra, R Pucket, K Arar, and K Hamby 2004 Real-time genotyping with oligonucleotide probes containing locked nucleic acids. Anal Biochem 324: 143-152.
Van Aerschot A, I Verheggen, C Hendrix,and P Herdewijn 1995 1,5-Anhydrohexitol nucleic acids, a new promising antisense construct. Angew Chem Int Ed 34: 1338–1339.
Tsuyoshi Yamamoto, Mariko Harada-Shiba, Moeka Nakatani, Shunsuke Wada, Hidenori Yasuhara, Keisuke Narukawa, Kiyomi Sasaki, Masa-Aki Shibata, Hidetaka Torigoe, Tetsuji Yamaoka, Takeshi Imanishi and Satoshi Obika; Cholesterol-lowering Action of BNA-based Antisense Oligonucleotides Targeting PCSK9 in Atherogenic Diet-induced Hypercholesterolemic Mice Molecular Therapy–Nucleic Acids (2012) 1, e22; oi:10.1038/mtna.2012.16.