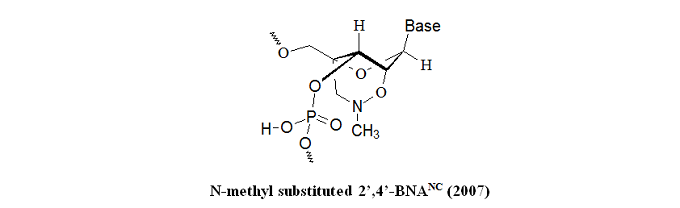
The research group synthesized a novel bridged nucleic acid 2’,4’-BNANC[N–Me] exhibiting high-affinity hybridization similar to that of 2’,4’-BNA (LNA) against an RNA complement. Furthermore, the scientists reported that the nucleic acid analog displayed RNA selectivity superior to 2’,4’-BNA (LNA) and other structural analogs of 2’,4’-BNA (LNA). Nuclease resistance of this nucleic acid analog is abundantly higher than that of 2’,4’-BNA (LNA) and slightly higher than that of a phosphorthioate. The hydrophobic methyl substituent on the backbone might present an additional advantage resulting in cellular uptake of the oligonucleotides. All of these reported characteristics of the BNA are essential for antisense applications. In the same year, Rahman et al. report that 2’,4’-BNANC form highly stable pyrimidine-motif DNA triplexes at physiological pH. These triplexes regulate gene expression, site-specific cleavage of DNA, gene mapping and isolation, maintenance of folded chromosome conformations, and gene-targeted mutagenesis. In a pyrimidine-motif triplex DNA, the triplex-forming oligonucleotide binds with the homopurine tract of the target duplex DNA in a sequence-specific manner through Hoogsteen hydrogen bonds to form T●A:T and C+●G:C triads. In the same year, Obika et al. report that 5’-amino-BNAs can be used to digest oligonucleotides triggered by triplex formation.
[2008] Imanishi’s group (Rahman et al. 2008) introduced three new bridged nucleic acid analogs called 2’,4’-BNANC[NH], 2’,4’-BNANC[NMe], and 2’,4’-BNANC[NBn]. The structures of these analogs are illustrated below. Rahman et al. considered the length of the bridged moiety during design. The research group designed a six-membered bridged structure with a unique structural feature (N-O bond) in the sugar moiety with a nitrogen atom. This atom can act as a conjugation site and improve the formation of duplexes and triplexes by lowering the repulsion between the negatively charged backbone phosphates.
Furthermore, the nitrogen atom on the bridge allows functionalization with hydrophobic and hydrophilic groups by adding bulky steric groups or any desired functional moiety. These modifications allow to control affinity towards complementary strands, regulate resistance against nuclease degradation, and synthesize active molecules designed for specific applications in genomics. The properties of these analogs were investigated and compared to those of previous 2’,4’-BNA (LNA) modified oligonucleotides. The chemical structures of the three 2’,4’-BNANC analogs are shown below.
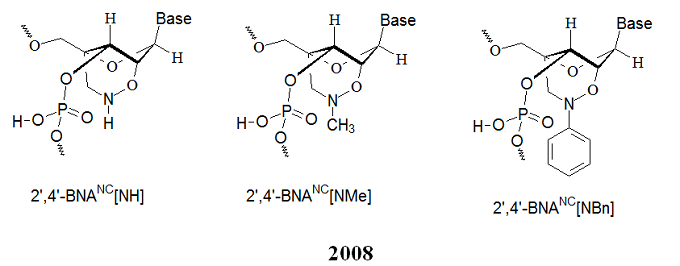
.
Compared to 2’,4’-BNA (LNA)-modified oligonucleotides, 2’,4’-BNANC congeners possess:
(i) Equal or higher binding affinity against an RNA complement with excellent single-mismatch discriminating power,
(ii) Much better RNA selective binding,
(iii) Stronger and more sequence selective triplex-forming characters, and
(iv) Immensely higher nuclease resistance, even higher than the Sp-phosphorthioate analog.
The researchers state that “2’,4’-BNANC-modified oligonucleotides with these excellent profiles show great promise for applications in antisense and antigene technologies.”
[2012] More recently, Yamamoto et al. demonstrated successfully that BNA-based antisense therapeutics inhibited hepatic PCSK9 expression, resulting in a substantial reduction of the serum LDL-C levels of mice. The findings support the hypothesis that PCSK9 is a potential therapeutic target for hypercholesterolemia. Apparently, this was the first time that researchers were able to show that BNA-based antisense oligonucleotides (AONs) induced cholesterol-lowering action in hypercholesterolemic mice. The study observed a moderate increase of aspartate aminotransferase, ALT, and blood urea nitrogen levels. However, the histopathological analysis revealed no severe hepatic toxicities. The same group, also in 2012, report that the 2’,4’-BNANC[NMe] analog, when used in antisense oligonucleotides, showed significantly stronger inhibitory activities, which is more pronounced in shorter (13- to 16mer) oligonucleotides. Their data led the researchers to conclude that the 2’,4’-BNANC[NMe] analog may be a better alternative to conventional LNAs.
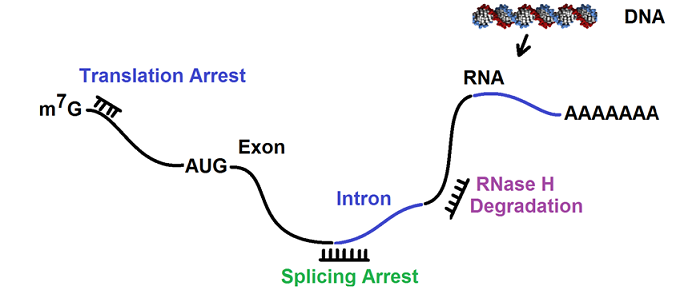
Action mechanism of antisense oligonucleotides
The proposed action mechanism for antisense oligonucleotides may involve translation arrest, mRNA degradation mediated by RNase H, and splicing arrest. The following figure illustrates the proposed action mechanism.
References
Ulla CHRISTENSEN, Nana JACOBSEN., Vivek K. RAJWANSHI, Jesper WENGEL and Troels KOCH. Stopped-flow kinetics of locked nucleic acid (LNA)–oligonucleotide duplex formation : studies of LNA–DNA and DNA–DNA interactions Biochem. J. (2001) 354, 481-484.
Yoshiyuki Hari, Satoshi Obika, Mitsuaki Sekiguchi and Takeshi Imanishi; Selective recognition of CG interruption by 2’,4’-BNA having 1-isoquinolone as a nucleobase in a pyrimidine motif triplex formationqTetrahedron 59 (2003) 5123–5128.
Makoto KOIZUMI; 2’-O,4’-C-Ethylene-Bridged Nucleic Acids (ENATM) as Next-Generation Antisense and Antigene Agents. Biol. Pharm. Bull. 27(4) 453-456 (2004).
Koizumi M.; ENA oligonucleotides as therapeutics. Curr Opin Mol Ther. 2006 Apr;8(2):144-9.
Koshkin AA, SK Singh, P Nielsen, VK Rajwanshi, R Kumar, M Meldgaard, CE Olsen, and J Wengel 1998 LNA (Locked Nucelic Acid): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 54: 3607-3630.
Koshkin, A.A., Nielsen, P., Meldgaard, M., Rajwanshi, V. K., Singh S. K. and Wengel, J. (1998). LNA (Locked Nucleic Acid): An RNA Mimic Forming Exceedingly Stable LNA:LNA Duplexes. J. Am. Chem. Soc. 120, 13252 – 13253.
Kazuyuki Miyashita, S. M. Abdur Rahman, Sayori Seki, Satoshi Obikaab and Takeshi Imanishi; N-Methyl substituted 2’,4’-BNANC: a highly nuclease-resistant nucleic acid analogue with high-affinity RNA selective hybridization. Chem. Commun., 2007, 3765–3767.
Koji Morita, Chikako Hasegawa, Masakatsu Kaneko, Shinya Tsutsum P, Junko Sone, Tomio Ishikawa, Takeshi Imanishi and Makoto Koizumi; 2'-O,4'-C-Ethylene-bridged nucleic acids (ENA) with nuclease resistance and high affinity for RNA. 2001. Nucleic Acids Research Supplement No. 1 241-242.
Nomenclature for polynucleotide chains including for the sugar puckering can be found at: http://www.chem.qmul.ac.uk/iupac/misc/pnuc2.html
Satoshi Obika, Daishu Nanbu, Yoshiyuki Hari, Ken.ichiro Morio, Yasuko In, Toshimasa Ishida, and Takeshi Imanishi; Synthesis of 2'-O,4'-C-Methyleneuridine and -cytidine. Novel Bicyclic Nucleosides Having a Fixed C3 ,-endo Sugar Puckering. Tetrahedron Letters, Vol. 38, No. 50, pp. 8735-8738, 1997.
Obika S, D Nanbu, Y Hari, J-i Andoh, K-i Morio, T Doi, and T Imanishi 1998. Stability and structural features of the duplexes containing nulcoeside analogs with a fixed N-type conformation. 2’-O, 4’-C methylene ribonucleosides. Tetrahedron Lett 39: 5401-5404.
Satoshi Obika, Mayumi Onoda, Koji Morita, Jun-ichi Andoh, Makoto Koizumi and Takeshi Imanishi; 3’-Amino-2’,4’-BNA: novel bridged nucleic acids having an N3’->P5’ phosphoramidate linkage. Chem. Commun., 2001, 1992–1993. Note: BNA/DNA; BNA/dsDNA.
Satoshi Obika, Yoshiyuki Hari, Mitsuaki Sekiguchi, and Takeshi Imanishi; A 2',4'-Bridged Nucleic Acid Containing 2-Pyridone as a Nucleobase: Efficient Recognition of a C●G Interruption by Triplex Formation with a Pyrimidine Motif. Angew. Chem. Int. Ed. 2001, 40, No. 11, 2079-2081.
Obika S, Hemamayi R, Masuda T, Sugimoto T, Nakagawa S, Mayumi T, Imanishi T.; Inhibition of ICAM-1 gene expression by antisense 2',4'-BNA oligonucleotides. Nucleic Acids Res Suppl. 2001;(1):145-6.
Satoshi Obika, Masaharu Tomizu, Yoshinori Negoro, Ayako Orita, Osamu Nakagawa, and Takeshi Imanishi; Double-Stranded DNA-Templated Oligonucleotide Digestion Triggered by Triplex Formation. ChemBioChem 2007, 8, 1924 – 1928. Note: Triplex triggered cleavage of oligonucleotides.
S. M. Abdur Rahman, Sayori Seki, Satoshi Obika, Sunao Haitani, Kazuyuki Miyashita, and Takeshi Imanishi; Highly Stable Pyrimidine-Motif Triplex Formation at Physiological pH Values by a Bridged Nucleic Acid Analogue. Angew. Chem. Int. Ed. 2007, 46, 4306 –4309.
Saenger, W.; Principles of Nucleic Acid Structure, Springer-Verlag, new York, 1984.
Eric E. Swayze, Andrew M. Siwkowski, Edward V. Wancewicz, Michael T. Migawa, Tadeusz K. Wyrzykiewicz, Gene Hung, Brett P. Monia, and and C. Frank Bennett; Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals Nucleic Acids Res. 2007 January; 35(2): 687–700.
Torigoe H, Y Hari, M Sekiguchi, S Obika, and T Imanishi 2001; 2’-O, 4’-C-methylene bridged nucleic acid modification promotes pyrimidine motif triplex DNA formation at physiologic pH. J Biol Chem 276: 2354-2360. Note: TFO for therapeutics.
Niels Tolstrup, Peter S. Nielsen, Jens G. Kolberg, Annett M. Frankel, Henrik Vissing, Sakari Kauppinen, OligoDesign: optimal design of LNA (locked nucleic acid) oligonucleotide capture probes for gene expression profiling, Nucleic Acids Research, Volume 31, Issue 13, 1 July 2003, Pages 3758–3762, https://doi.org/10.1093/nar/gkg580 , https://academic.oup.com/nar/article/31/13/3758/2904202?login=true
Tsuyoshi Yamamoto, Mariko Harada-Shiba, Moeka Nakatani, Shunsuke Wada, Hidenori Yasuhara, Keisuke Narukawa, Kiyomi Sasaki, Masa-Aki Shibata, Hidetaka Torigoe, Tetsuji Yamaoka, Takeshi Imanishi and Satoshi Obika; Cholesterol-lowering Action of BNA-based Antisense Oligonucleotides Targeting PCSK9 in Atherogenic Diet-induced Hypercholesterolemic Mice Molecular Therapy–Nucleic Acids (2012) 1, e22; oi:10.1038/mtna.2012.16.
Tsuyoshi Yamamoto, Hidenori Yasuhara, FumitoWada, Mariko Harada-Shiba, Takeshi Imanishi, and Satoshi Obika; Superior Silencing by 2’,4’-BNANC-Based Short Antisense Oligonucleotides Compared to 2’,4’-BNA/LNA-Based Apolipoprotein B Antisense Inhibitors. Hindawi Publishing Corporation, Journal of Nucleic Acids Volume 2012, Article ID 707323, 7 pages doi:10.1155/2012/707323.
Yong You, Bernardo G. Moreira, Mark A. Behlke, and Richard Owczarzy; Design of LNA probes that improve mismatch discrimination. Nucleic Acids Research, 2006, Vol. 34, No. 8 e60.
--...--