Mycobacterium tuberculosis Peptide Libraries
Design of Mtb Peptide Libraries for Targeted Research
Tuberculosis has become an escalating global health problem in recent years. It is estimated that one-third of the world’s population presently is infected with Mycobacterium tuberculosis (Mtb) and that the bacteria causes 1.7 million deaths annually. On November 20th, 2012, the news reported that students in Grand Forks, ND, were diagnosed with tuberculosis and health officials reported three new cases of tuberculosis (TB) in Grand Forks, including a student at Valley Middle School. This brought the total number of active cases to 13. An article in the Wall Street Journal on Friday November 23, 2012 reported how the fight against TB made the bacteria stronger.The standard treatment to cure TB in India and other countries with a high infection rate to give patients a cocktail of antibiotic drugs at a cost of $9 per month without prior testing for drug-resistant strains. This standard treatment allowed the bacteria to evolve more resistant strains. Additional to the cost for testing in a certified lab to help guide a successful treatment, the cost to treat resistant TB strains amounts to $2,000 per month. Chances are that until an integrated approach that uses testing for drug-resistance together with a targeted treatment is established, the infection rate of TB will continue to increase worldwide.
TB is a bacterial infection caused by the bacteria Mycobacterium tuberculosis which usually attacks the lungs, but can also spread to other parts of the body. TB disperses into the air when a person with TB of the lungs or throat coughs, sneezes or talks. Only a medical examination will tell if a person has been exposed to the bacteria. People with weak immune systems have a higher risk for infection. Symptoms of TB in the lungs may include a bad cough that lasts three weeks or longer, weight loss, coughing up blood or mucus, weakness or fatigue, fever and chills, and night sweats. TB can be deadly if not treated properly and it is highly recommend that people experiencing these symptoms go to a doctor immediately. Exposure to tuberculosis that includes frequent or prolonged exposure, such as sitting in a small room or in a confined area for a long period of time with someone who has active TB, is infectious. However, people are only contagious when there is active disease in their lungs or throat that has not been treated. TB does not spread through clothes, dishes, floors or furniture. The estimate is that in India the disease kills 300,000 people out of 1.2 billion a year. Unfortunately, anyone can get TB. Individuals with weakened immune systems, including those with AIDS or those infected with HIV, have a higher risk to get infected. Active TB can usually be cured with several medicines taken for a long period of time. Latent TB can be treated to prevent the development of active TB. The standard tuberculosis treatment is to take a combination of antibiotics for half a year or more. If patients quit the therapy prematurely, the risk that drug-resistant strains emerge increases. Mycobacteria are hard to kill and it was assumed that dormant cells less susceptible to antibiotics exist even in patients with active disease. Recently, a study discovered that Mycobacteria divide asymmetrically. This behavior generates a population of cells that grow at different rates, have different sizes, and differ in how susceptible they are to antibiotics. This gives the bacteria an increased chance for survival. These new findings may aid in the development of new drugs against the hard to kill Mtb cells.
The origin of the Mtb complex, its spread and demography was studied in 2008 by Wirth et al. using a new approach that employed genetic markers based on mycobacterial interspersed repetitive units. These genetic loci comprise variable numbers and tandem repeat sequences that allowed them to be used as genotyping markers. Similar to microsatellites, they behave as selectively neutral phylogenetic markers if large numbers of loci are used. The research group used these markers to calculate a molecular clock and to model their evolution. They report that the Mtb complex consists of two independent clades. One composed of lineages from humans and a second composed of lineages from both animal and human isolates. Furthermore, the results provide genetic evidence that the most common ancestor for this bacterial complex emerged some 40,000 years ago from its progenitor in East Africa, where it spread in parallel to human migration routes.
The World Health Organization reports on their web site that “tuberculosis (TB) is second only to HIV/AIDS as the greatest killer worldwide due to a single infectious agent. In 2011, 8.7 million people fell ill with TB and 1.4 million died from TB. Over 95% of TB deaths occur in low- and middle-income countries. TB is among the top three causes of death for women aged 15 to 44. In 2010, there were about 10 million orphan children as a result of TB deaths among parents. TB is a leading killer of people living with HIV causing one quarter of all deaths. Multi-drug resistant TB (MDR-TB) is present in virtually all countries surveyed.” The good news is that the estimated number of people falling ill with tuberculosis each year is slowly declining. The TB death rate dropped 41% between 1990 and 2011. TB occurs in every part of the world. In 2011, the largest number of new TB cases occurred in Asia, accounting for 60% of new cases globally. Sub-Saharan Africa carried the greatest proportion of new cases per population with over 260 cases per 100 000 population in 2011.
Mtb Peptide Libraries Can be Used to Help Developing New Vaccines Against TB
There is an urgent need to design new antituberculosis vaccines. The anti-TB vaccine presently available has a high variability and is ineffective in adults. The goal for immunologist is to design a vaccine that confers complete protection against the disease. Presently, research is focused on finding new Mtb-specific antigens that can be used to replace or improve the old vaccine. Early innate and adaptive responses to Mtb are critical for the successful containment of the infection. However, these events are poorly understood, in part because low-dose aerogenic inoculation does not trigger a robust early inflammatory response. Complicating this is the fact that very early T-cell responses are difficult to study. There remains many open questions such as if Mtb delays the onset of early immunity, either by accident or design, and if true memory T lymphocytes can be elicited by vaccination. New research tools are now available to help improve our understanding of TB immunity. Studying the host genetic factors such as human leucocyte antigens (HLA) and non-HLA genes/gene products that are associated with the susceptibility to TB has the potential to determine genetic markers that help in understanding predisposition factors that allow for the development of the disease. Establishing a clear picture of the immune response network to this pathogen is essential for the design of effective vaccines. Identification of high-activity binding peptides that are able to inhibit bacterial invasion is a step toward this goal. Ocampo et al. in 2012 report the identification of Mtb Rv3166c protein high-activity binding peptides (HABPs) that were able to inhibit bacterial invasion of U937 (monocyte-derived macrophages) and A549 (type II alveolar epithelial cells) cell lines. The researchers used PCR to confirm the presence and transcription of the rv3166c gene in the Mtb species complex. Western blotting and immunoelectron microscopy was used to evaluate and confirm Rv3166c expression which was found to be present mainly on the cell surface. The research group synthesized sixteen 21mer peptides that covered the entire length of the protein and contained a tyrosine on the C-terminal end. The tyrosine allowed the radiolabeling of the peptides after synthesis using I125 to be used in a binding assay. The peptides were tested for their ability to bind to U937 and A549 cells. Two U937 HABPs were identified and three for A549, one of them being shared by both cell lines. Four peptides were identified to inhibited Mtb entry by 15.07–94.06%. These results led the researchers to conclude that Rv3166c HABPs can be used as candidates for further studies contributing towards the search for a multiepitope, chemically synthesized, subunit-based antituberculosis vaccine. The next table lists the peptides used for the study.
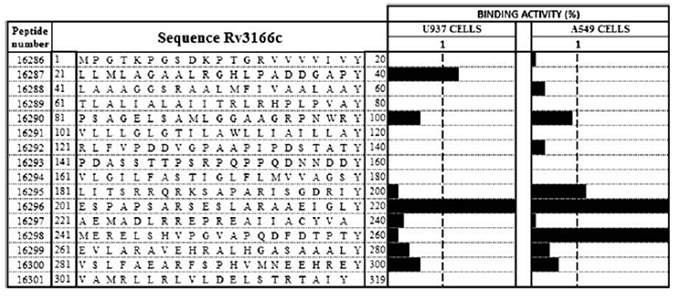
The graph shows the Rv3166c peptide binding activity profile. The amino acid sequences and specific U937 and A549 cell binding activities for Rv3166c peptides are listed. Tyr was added at the C-terminal end of those peptides lacking it to facilitate I125 radiolabeling. The black bars on the right-hand side represent each peptide’s binding activity determined as the specific binding/total peptide added ratio. The dotted line separates peptides having ≥1% binding activity (Ocampo et al.).
The key to generating good experimental results is the design of the experimental approach on how to use scientific tools such as peptide libraries. Below is the description of the design of peptide libraries or pools using a few selected examples.
Example of Peptide Library Design Using the Sequence of the Hypothetical Protein Rv3166c [Mycobacterium tuberculosis H37Rv] NP_217682.1:
1. Non-Overlapping Peptide Libraries:
This example shows the design of a non-overlapping library. First, select the sequences of peptides to be synthesized. In this case, 20mer peptides selected are highlighted in alternating grey shadowing.
The library for this selected protein consists of sixteen peptides. The selected peptides can be modified either on the N- or C-terminal end with tyrosine to allow for the use of I125 as the radiolabel, cysteine, to allow for conjugation to another compound, beads, or biotin. The modifications also allow binding to avidin or streptavidin coated beads, or any other desired label such as a fluorophore or stable isotope tag, which allow for the coding of the peptides. The use for the downstream analysis tools will determine what type of label needs to be selected.
2. Overlapping Peptide Libraries:
The next example shows the design of a library that uses 20mer peptides and an overlap of eleven amino acid residues. This type of peptide library can be designed by using the whole sequence of the original protein or protein domain to generate many equal-length overlapping peptide fragments. Typical applications are continuous epitope mapping and T-cell epitope determination. The “Peptide Library Tools” that can be found on Bio-Synthesis Inc.’s web site was used for the design. The following link shows the location of the tool: http://www.biosyn.com/PeptideDesignLibrary/PeptideDesignLibrary.aspx.
The resulting list of selected peptides is shown in the following table.
Many more permutations of peptide library designs are possible. The resulting libraries can be used to screen highly active compounds such as antigenic peptides, receptor ligands, antimicrobial compounds, protein binding interfaces, and enzyme inhibitors, among others.
Major applications are: the use of peptide libraries for epitope mapping studies, vaccine research, high-throughput protein interaction analysis, customized peptide microarray production and kinase assays. Epitope mapping requires the design of overlapping peptide libraries, which can be customized by adjusting the fragment length and offset number for the optimum balance between low costs and high data value.
Next, the different library types are explained.
Alanine Peptide Scanning Library: The design of a peptide library in which alanine (Ala, A) is systematically substituted into each of the amino acids which can be used to identify epitope activity. This is sometimes also called an Alanine Scanning Library.
Positional Peptide Library: A selected position in a peptide sequence is systematically replaced with a different amino acid to study the effect of the substituted amino acid at a certain position.
Truncation Peptide Library: A truncation library can be used to predict the minimum amino acid sequence length required for optimum epitope activity.
Random Peptide Library: To design this library, selected positions within the original peptide sequence are randomly substituting other natural amino acids in a shot-gun type approach help find potential alternative sequences for enhanced peptide activity.
Scramble Peptide Library: A scrambled library is designed by carrying out permutation on the original peptide's sequence. It has the potential to give all possible alternatives and offers and represents the highest degree of variability per peptide library.
T-cell Truncated Peptide Libraries: This library allows the testing of all possible T-cell epitopes across a protein of interest.
Examples of Mtb Peptide Libraries and Their Use Found in the Literature
Kovjazin in 2010 reported the use of bioinformatic methods to find Mtb peptides with high epitope densities that can be used for vaccine design. Signal peptides and trans-membrane domains were found to have exceptionally high epitope densities. The researchers explain that the major histocompatibility complex (MHC)-binding of these domains relies on their hydrophobic nature and their specific sequence. The research group computed the epitope densities for signal peptides and experimentally confirmed their immunogenicity using a panel of nine synthetic peptides in vitro. The scientists reason that the high epitope density may be the result of an arbitrary overlap between the typical patterns of signal peptides and frequent MHC binding motifs, and that the high epitope density in such domains could be explained on the basis of an evolutionary advantage caused by preferential presentation of hydrophobic sequences especially for the signal peptides. It is known that host-pathogen, coevolutionary pressures can change the relative frequency of the different HLA alleles in a population and it is thus possible that HLA alleles preferentially presenting signal peptide epitopes were selected. Highly conserved signal peptide motifs are almost similar in Eukaryotes and Prokaryotes. The authors reason that these results may indicate that pathogens use secreted proteins to manipulate their hosts, and that the host has evolved a mechanisms to present bacterial peptides on MHC-I molecules.
Joosten et al. in 2010 describe responses of human T-cells to Mtb-derived peptides containing predicted HLA-E binding motifs. A total of 69 peptides were used for the study. The CD8+ T-cell is one player of the immune response against Mtb. CD8+ T-cells recognize infected cells through Mtb derived peptides that are presented on HLA class I molecules. The nonclassical HLA molecule HLA-E was studied as a presenter of Mtb antigens. The Mtb genome contains multiple sequences that can be presented by human HLA-E. These peptides were recognized by CD8+ T-cells from healthy individuals that were sensitized to Mtb, resulting in CD8+ T-cell proliferation. The investigated T-cells were able to lyse mycobacterium infected cells in a HLA-E restricted manner. Additionally, these T-cells also inhibited proliferation of other T-cells in their vicinity, which is a property of regulatory T-cells.
Dye and Williams report in 2010 that M. tuberculosis is a family of strains and lineages and that the TB epidemiology is currently changing. Recent investigations have revealed hypervariable regions in the genome, the African origins, he spread around the world of the pathogen, and which genes code for drug resistance. The data demonstrated that different strains spread more or less quickly through populations and are more or less capable to cause active TB. This indicates that the magnitude of the variations and its epidemiological consequences are mostly unquantified. The analysis of different strains to determine their genetics and phenotypic differences may be needed to allow for a better control of the disease in the future.
|
Russel et al in 2010 described the life cycle of M. Tuberculosis in the journal Science. They state that, “There is no effective vaccine against infection, and current drug therapies are fraught with problems, predominantly because of the protracted nature of the treatment and the increasing occurrence of drug resistance.” The life cycle of M. tuberculosis (see figure) begins when Mtb bacilli, present in exhaled droplets or nuclei, are inhaled, engulfed and digested by resident alveolar macrophages, they initiate the infection. The result is a pro-inflammatory response which triggers the infected cells to invade the epithelial tissue. Monocytes are recruited from the circulation and extensive neovascularization occurs in the infection site. Neovascularization refers to the formation of functional microvascular networks with red blood cell perfusion. The macrophages in the granulomas differentiate to form epithelioid cells, multinucleate giant cells, and foam cells filled with lipid droplets. The granuloma, a tiny collection of
|
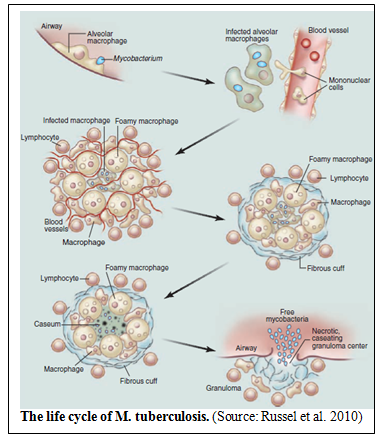 |
immune cells known as macrophages, can become further stratified by the formation of a fibrous extra layer of extracellular matrix material that is laid down outside the macrophage layer. Lymphocytes appear to be restricted primarily to this peripheral area. Many of the granulomas persist in this balanced state, but progression toward disease is characterized by the loss of vascularization, increased necrosis, and the accumulation of caseum in the granuloma center. Ultimately, infectious bacilli are released into the airways when the granuloma cavitates and collapses into the lungs (Russel at al. 2010). According to the paper, the research field is lacking some of the most basic tools to evaluate or assess the beneficial effect of new drugs. Furthermore, no clear biomarkers to assess the disease status are presently available.
Chun et al. in 2001 report the use of N-formylated Mtb peptides to test their ability to bind M3 using an immunofluorescence-based peptide-binding assay. M3 is an MHC class Ib molecule that preferentially presents N-formulated peptides to CD8+ T cells. Bacteria initiate protein synthesis with N-formylated methionine, which makes M3 especially suitable for presenting this type of peptide epitopes. Therefore, the research group scanned the full sequence of the Mtb genome for N-terminal peptides that shared common features with other M3-binding peptides using bioinformatic tools. Synthetic peptides corresponding to the selected sequences were tested for their ability to bind M3. Furthermore, the researchers report that four of these peptides were able to elicit cytotoxic T lymphocytes (CTLs) from mice immunized with peptide-coated splenocytes. They concluded that their data suggest that M3-restricted T cells may participate in the immune response to Mtb.
References
Taehoon Chun, Natalya V. Serbina, Dawn Nolt, Bin Wang, Nancy M. Chiu, JoAnne L. Flynn, and Chyung-Ru Wang; Induction of M3-restricted Cytotoxic T Lymphocyte Responses by N-formylated Peptides Derived from Mycobacterium tuberculosis. J. Exp. Med. Volume 193, Number 10, May 21, 2001 1213–1220.
Christopher Dye and Brian G. Williams; The Population Dynamics and Control of Tuberculosis. Science 328, 856 (2010).
Simone A. Joosten, Krista E. van Meijgaarden, Pascale C. van Weeren, Fatima Kazi, Annemieke Geluk, Nigel D. L. Savage; Mycobacterium tuberculosis Peptides Presented by HLA-E Molecules Are Targets for Human CD8+ T-Cells with Cytotoxic as well as Regulatory Activity. PLoS Pathogens | www.plospathogens.org 1 February 2010 | Volume 6 | Issue 2 | e1000782.
Riva Kovjazina, Ilan Volovitzc, Yair Daona, Tal ViderShalitb, Roy Azranb, Lea Tsabanb, Lior Carmona, Yoram Louzoun; Signal peptides and transmembrane regions are broadly immunogenic and have high CD8+ T cell epitope densities: Implications for vaccine development. Molecular Immunology 48 (2011) 1009–1018.
Frieder M, Lewinsohn DM. T-cell epitope mapping in Mycobacterium tuberculosis using pepmixes created by micro-scale SPOT- synthesis. Methods Mol Biol. 2009;524:369-82.
Marisol Ocampo, Daniel Aristiza´bal-Ramı´rez, Diana M.Rodrı´guez, Marina Mun˜oz, Hernando Curtidor, Magnolia Vanegas, Manuel A.Patarroyo, and Manuel E.Patarroyo; The role of Mycobacterium tuberculosis Rv3166c protein-derived high-activity binding peptides in inhibiting invasion of human cell lines. Protein Engineering, Design & Selection vol. 25 no. 5 pp. 235–242, 2012.
David G. Russell, Clifton E. Barry 3rd, JoAnne L. Flynn; Tuberculosis: What We Don't Know Can, and Does, Hurt Us. Science 328, 852 (2010).
Winslow GM, Cooper A, Reiley W, Chatterjee M, Woodland DL; Early T-cell responses in tuberculosis immunity. Immunol Rev. 2008 Oct; 225:284-99.
Wirth T, Hildebrand F, Allix-Béguec C, Wölbeling F, Kubica T, et al. (2008) Origin, Spread and Demography of the Mycobacterium tuberculosis Complex. PLoS Pathog 4(9): e1000160. doi:10.1371/journal.ppat.1000160