Purines, Pyrimidines, and Nucleotides
Purines, pyrimidines, and nucleotides are ubiquitous molecules found throughout a mammalian as well as a human body. In one form or another, these molecules serve a variety of roles. Nucleotides are molecular building blocks or subunits of nucleic acids such as deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). These subunits are also called monomers. The synthesis of purines, pyrimidines, and nucleotides is an important part of mammalian metabolism. Errors in purine and pyrimidine synthesis and metabolism, inborn or acquired, often are the cause of disease or ultimately lead to disease.
Nucleotides and their structure
A nucleotide is made up of three units:
- A nitrogen-containing base,
- A five-carbon sugar,
- One, two or three phosphate groups.
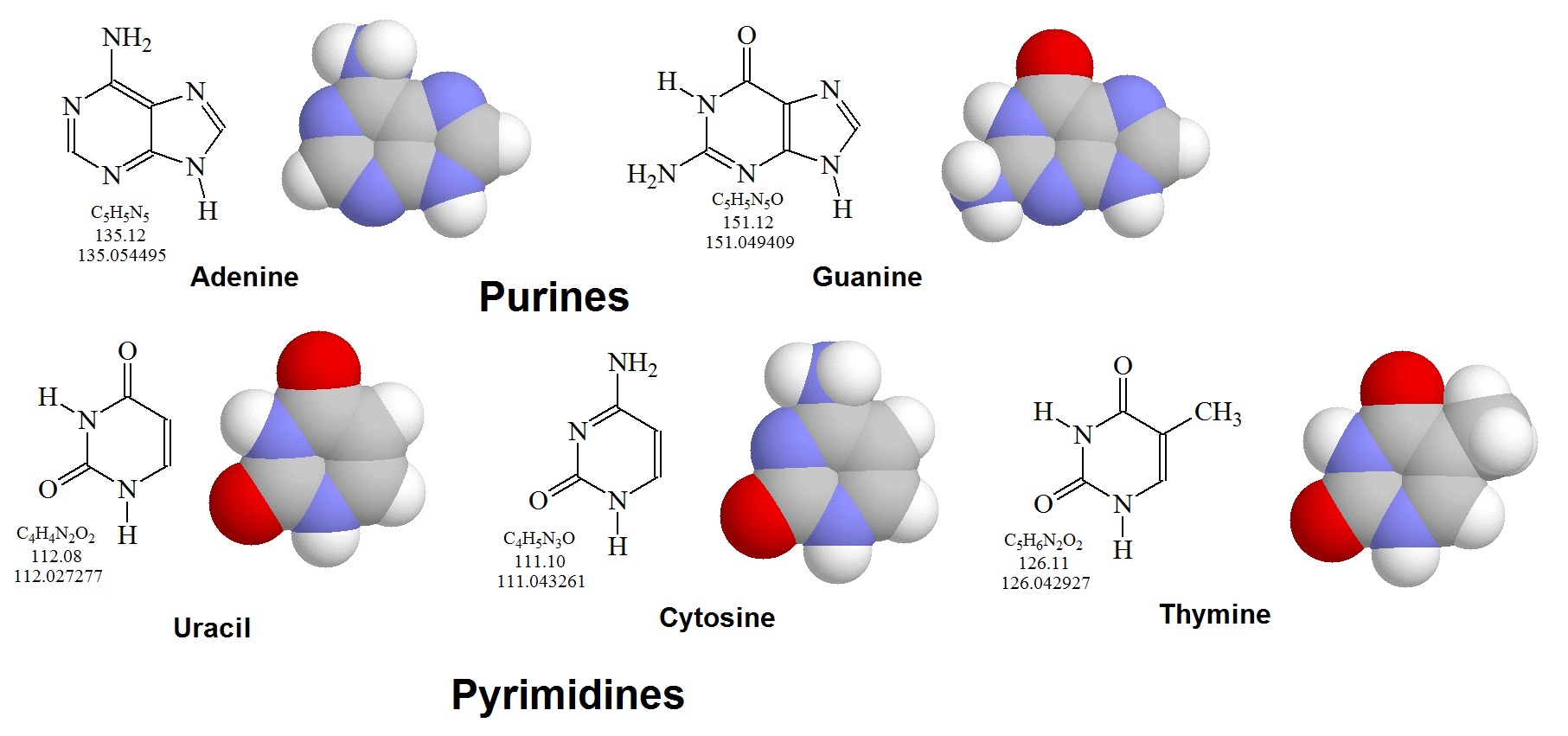
There are five common bases. Two of these bases are derivatives of purine. These are adenine and guanine. Three of the bases are pyrimidine derivatives. These are cytosine, thymine, and uracil. The addition of ribose moiety or 2-deoxyribose moiety to a base forms a nucleoside. The figure below illustrates the general structures of these molecules. Adenine, guanine, and cytosine can be found bonded to either a ribose or a 2-deoxyribose molecule. Uracil is usually found in combination with a ribose. Thymine is usually bonded to 2-deoxyribose. Ribonucleosides are called adenosine, guanosine, cytidine, and uridine. Deoxyribonucleotides are called deoxyadenosine, deoxyguanosine, deoxycytidine, and deoxythymidine.
.jpg)
Figure 1: General nucleotide structure showing the numbering convention for the pentose ring. The carbon atoms of the pentoses are numbered with primes.
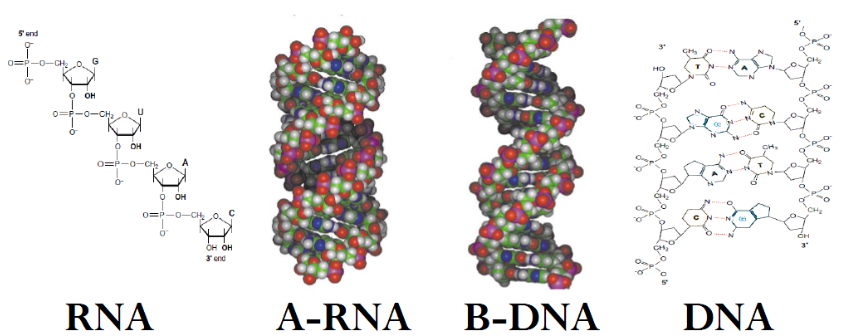
Figure 2: Helical structures of A-RNA (1RNA) and B-DNA (3BSE). The N-type sugar puckering exists predominently in duplexes of A-RNA and the S-type in duplexes with B-DNA helical structure. Models of the structures were produced using Pymole using X-ray structure data downloaded from the RCSB Protein Structure Data Bank.
IUPAC nucleotide codes with ambiguity
Reference:
Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Research. 1985;13(9):3021-3030.
IUPAC Standards: INTERNATIONAL UNION OF PURE AND APPLIED CHEMISTRY AND INTERNATIONAL UNION OF BIOCHEMISTRY ABBREVIATIONS AND SYMBOLS FOR NUCLEIC ACIDS, POLYNUCLEOTIDES AND THEIR CONSTITUENTS RULES APPROVED 1974 Issued by the JUPAC—IUB Commission on Biochemical Nomenclature
Johnson, Andrew D.; An extended IUPAC nomenclature code for polymorphic nucleic acids. Bioinformatics. 2010 May 15; 26(10): 1386–1389.