All newly formed eukaryotic RNA transcripts undergo a number of post-translational modifications before they are transferred into the cytoplasm. Eukaryotic mRNAs are initially transcribed by RNA polymerase II as direct copies of a gene that include several non-coding sequences such as introns. A nuclear RNA molecule experiences three main events during its life:
- The addition of a 5’ cap,
- The addition of a 3’ tail,
- The removal of unwanted intronic sequences by a process called RNA splicing.
Splicing is a particularly complex process and some eukaryotic cells can make different proteins by forming different mRNAs from the same gene, also called alternative splicing. In addition, the identification of post-translational modifications within mRNA molecules has been quite important in the development of in vivo therapeutic strategies, such as the anti-sense oligonuleotides based technology, and in vitro gene manipulations, such as the construction of cDNA libraries.
During transcription eukaryotic RNA becomes associated with small ribonuclear proteins that add a cap structure to the 5’ end. The majority of eukaryotic and viral messenger RNAs apparently contain a cap structure. RNA molecules that are 20 to 30 nucleotides long are capped by the initial addition of a guanine nucleotide to the first RNA base by a reaction catalyzed by the enzyme guanalyl transferase.
Transcription starts with a nucleotide triphosphate which is usually a purine (A or G). The first nucleotide has a 5’ triphosphate group and the usual phosphodiester bond on its 3’ position which is connected to the 5’ position of the next nucleotide. The initial sequence is: 5’ ppp(A or G)pNpNpNp… .
The 5’ 7-methylguanosine (m7G) cap is a unique feature of eukaryotic cellular and viral mRNA and small nuclear and nucleolar RNAs involved in nuclear RNA processing reactions. Capping appears to be essential for eukaryotic cells. The cap is attached backwards by the formation of a 5’ to 5’ linkage of the first RNA base. The N7 atom of the guanine is also methylated following the addition. This reaction generates a 7-methyl guanine and this cap structure is also called a “cap 0” structure. Furthermore, the adjacent ribose units can also be methylated at the C2 position.
If the C2 position at the first ribose is methylated the cap is called “cap 1” structure, and if the C2 positions on both riboses are methylated it is called a “cap 2” structure. The ‘cap O’ structure, N7-methylguanosine-triphosphate (7mGpppN), at the 5′-terminus is required in varying degrees for processing and maturation of the RNA transcript in the nucleus. The precursor to the N7-MeGppp cap is the unmethylated version, 5′ guanosine-triphosphate (Gppp).
The cap is formed by three enzymatic reactions at the 5’ terminus of the nascent mRNAs. Structures of the different m7G caps and of the precursor of the N7-MeGppp cap which is the unmethylated version, 5′ guanosine-triphosphate (Gppp), are shown below.
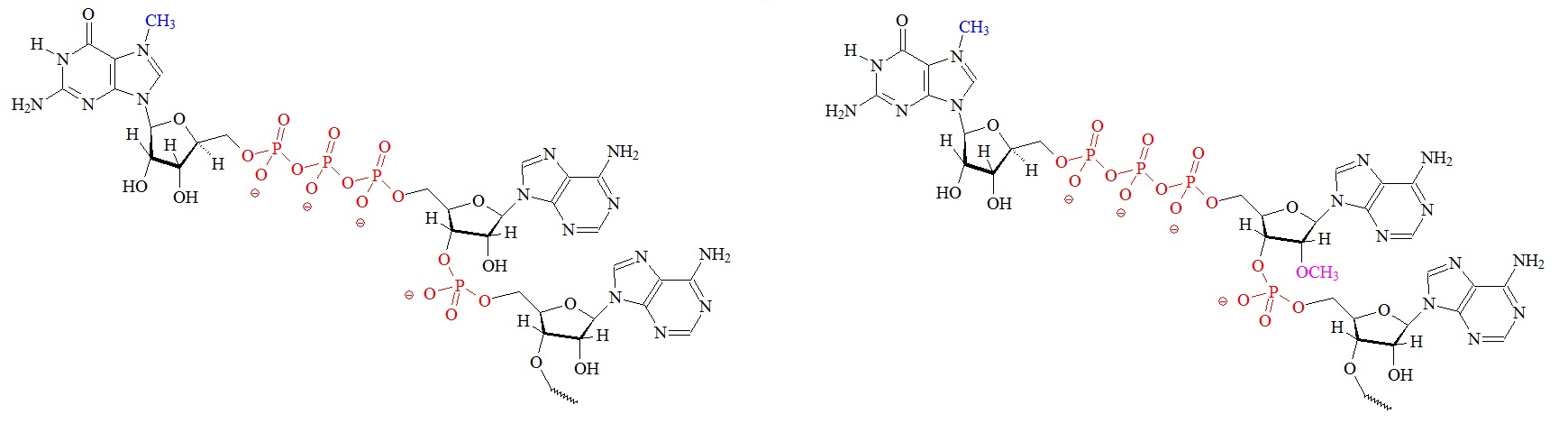
A) m7G cap “cap O” structure (7-methyl guanine) B) “cap 1” structure; 2’ mono methylated
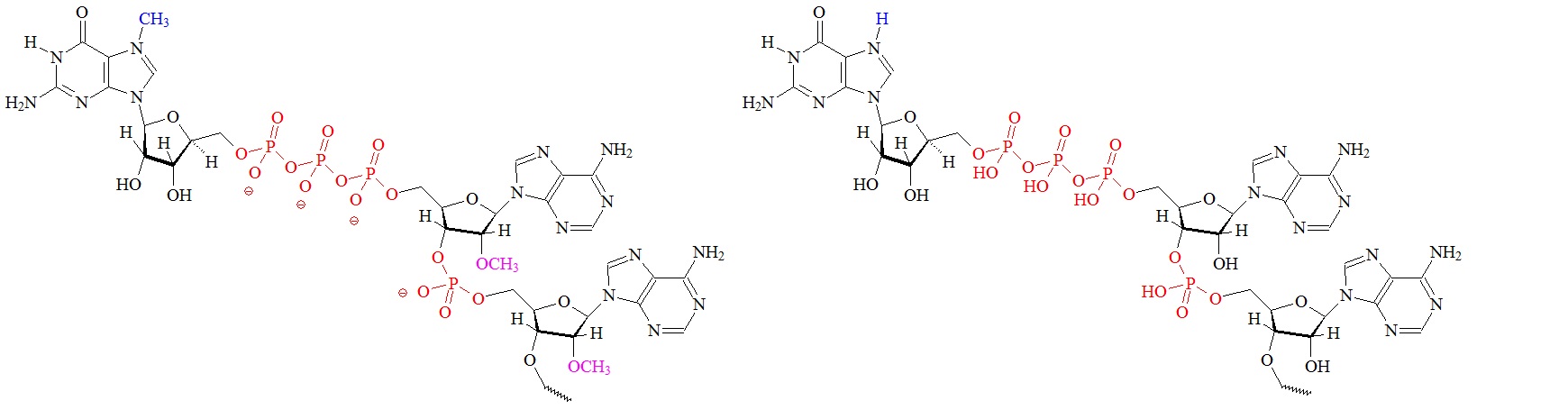
C) “cap 2” structure; 2’ dimethylated D) Precursor for the m7G cap
Caps are believed to serve several functions. They may be (i) involved in subsequent splicing reactions, (ii) and/or serve as positioning guides for translation. The observation that ribosomes can not bind to uncapped messages supports this notion. Furthermore, capping may also determine the stability of transcripts explaining observed differences in the half-lives between prokaryotic and eukaryotic mRNAs. In addition, the observation that cap analogs can inhibit splicing when added at the start of the reaction but not at later times of incubation suggests that the cap recognition is an important step in the formation of a specific ribonucleoprotein complex required for splicing. Several studies including X-ray crystallography have now shown that the cap structure is necessary for optimal mRNA translation, that it participates in the splicing of mRNA precursors, and affects nuclear export and mRNA stability. Furthermore, the cap function in translation is mediated by the 25 kDa protein eukaryotic initiation factor 4E (eIF4E).
Different forms of caps, for example, m7GpppN on mRNAs and m2'7GpppN on low-molecular-weight nuclear RNAs, may interact with proteins that may be needed for the generation of mature cellular mRNAs. Studies using affinity photolabeling methods indicated that the cap binding activity in influenza virus is associated with a P protein.
The next figure shows a summary of the possible different modifications on the 5’ guanosine-triphosphate cap.
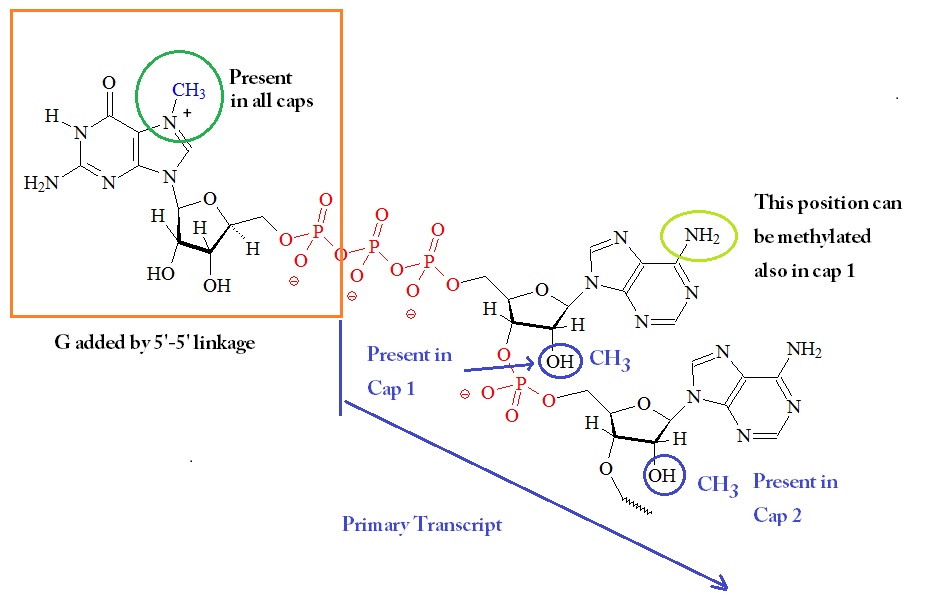
5'-Guanosine-triphosphate Cap
The cap blocks the 5’ end of mRNA and may be methylated at several positions.
Synthesis and purification of caps and capped structures
Capped RNA oligonucleotides can be synthesized enzymatically by in vitro transcription and chemically using phosphoramidite chemistries. The use of chemical synthesis allows reproducibly obtaining capped RNAs at almost any scale independent from the RNA sequences and allows the addition of modifications at specific positions as well. The resulting modified oligonucleotides can be purified to a high purity and analyzed by a number of established methods, including PAGE, reverse phase HPLC, anion-exchange HPLC and mass-spectrometry. Since free 2’, 3’-hydroxyls can form complexes with borate ions, caps or capped oligonucleotides can be purified by affinity chromatography using substrates such as dihydroxyboryl-cellulose. Treatment with periodate allows to oxidize the cis-diols to dialdehyde which can then be reduced with 3H borohydrate to label the terminus radioactively. Alternatively, fluorophores may be added using this chemistry as well.
References
Hakansson K, Doherty AJ, Shuman S, Wigley DB; X-ray crystallography reveals a large conformational change during guanyl transfer by mrna capping enzymes. Cell(Cambridge,Mass.) (1997) 89 p.545. [PubMed]
Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991 Oct 25;266(30):19867-70. [PubMed]
HANS KROATH AND AARON J. SHATKIN; mRNA 5'-Cap Binding Activity in Purified Influenza Virus. JOURNAL OF VIROLOGY, Mar. 1982, p. 1105-1108 Vol. 41, No. 3. [PMC]
O'Mullane L, Eperon IC. The pre-mRNA 5' cap determines whether U6 small nuclear RNA succeeds U1 small nuclear ribonucleoprotein particle at 5' splice sites. Mol Cell Biol. 1998 Dec;18(12):7510-20. [PMC]
Lewin, Benjamin; Genes IV. Oxford University Press 1990. [Book]
Anna Niedzwiecka, Joseph Marcotrigiano, Janusz Stepinski, Marzena Jankowska-Anyszka, Aleksandra Wyslouch-Cieszynska, Michal Dadlez, Anne-Claude Gingras, Pawel Mak, Edward Darzynkiewicz, Nahum Sonenberg, Stephen K. Burley and Ryszard Stolarski; Biophysical Studies of eIF4E Cap-binding Protein: Recognition of mRNA 50 Cap Structure and Synthetic Fragments of eIF4G and 4E-BP1 Proteins. J. Mol. Biol. (2002) 319, 615–635. [PubMed]
Shatkin AJ. Capping of eucaryotic mRNAs. Cell. 1976 Dec;9(4 PT 2):645-53. [PubMed]
Shih DS, Dasgupta R, Kaesberg P. 7-Methyl-guanosine and efficiency of RNA translation. J Virol. 1976 Aug;19(2):637-42. [PMC]
Stewart Shuman; What messenger RNA capping tells us about eukaryotic evolution. Nature Reviews Molecular Cell Biology 3, 619-625 (August 2002). [PubMed]
---...---