Bridged Nucleic Acids (BNAs) can be used as tools for DNA or RNA targeting!
Synthetic oligonucleotides have emerged as imported and established tools in the life sciences enabling many applications in biology, genetics, diagnostics, molecular biology and molecular medicine, as well as in other scientific fields. Natural oligodeoxynucleotides can form DNA:DNA and DNA:RNA duplexes but are often unstable and labile to nucleases. As a result, different nucleic acid analogs have been designed and synthesized for the enhancement of high-affinity recognition of DNA and RNA targets, duplex stability and to assist with cellular uptake. Bridged Nucleic Acids (BNAs) are a good example for these.
Bridged Nucleic Acids (BNAs) contain either a five-membered or six-membered bridged structure. BNAs contain a synthetically incorporated bridge at the 2’, 4’-position of the ribose to afford a 2’, 4’-BNA monomer. Monomers can be incorporated into oligonucleotide polymer structures using standard phosphoramidite chemistry.
BNAs oligonucleotide mimics exhibit
(i) Equal or higher binding affinities against RNA complements as well as
excellent single-mismatch discriminating power,
(ii) Much better RNA selective binding,
(iii) Stronger and more sequence selective triplex-forming characters, and
(iv) A pronounced higher nuclease resistance, even higher than
Sp-phosphorthioate analogs, or than regular DNA, or RNA
oligonucleotides.
2',4-BNANC [NMe]
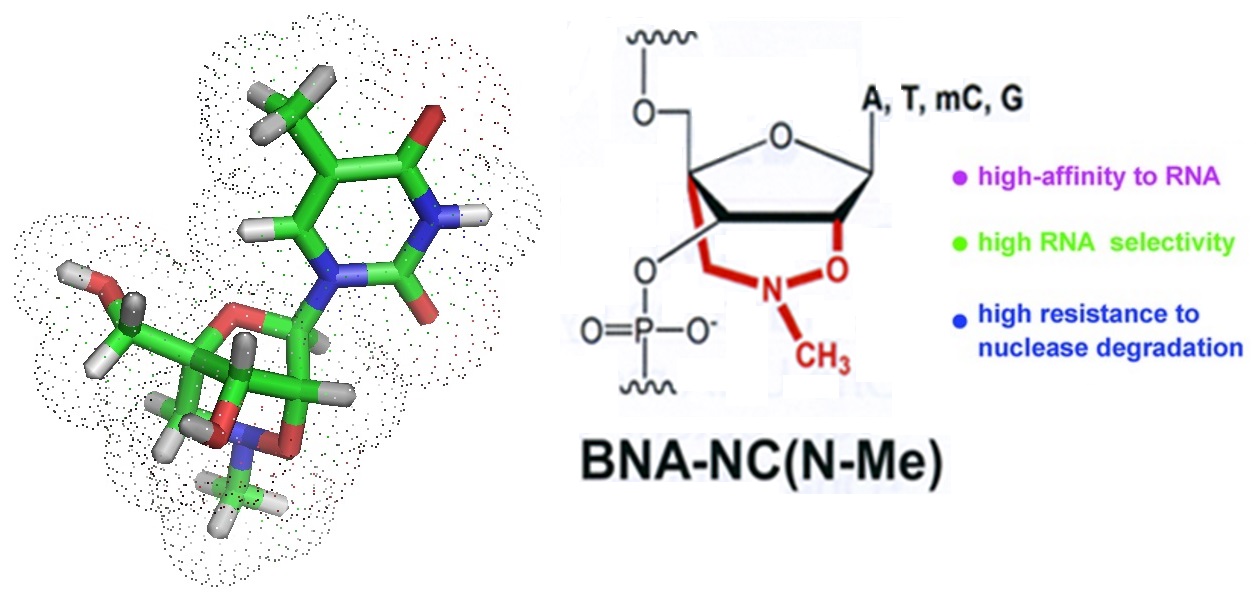
BNA 2’, 4’-BNANC [NMe] has a six-membered bridged ring structure with a unique structural feature, an N-O bond, which is a hydrophilic amino-oxy moiety, wihin the sugar moiety. A methyl group is attached to the nitrogen atom located in the bridge. Oligonucleotides modified with this BNA have a very high affinity to RNA and are resistant to endonucleolytic cleavage by nucleases. Furthermore, 2’, 4’-BNANC [NMe]s are less toxic to hepatic cells than other bridged nucleic acids, for example LNAs. Bridged nucleic acids (BNAs) are very useful tools for DNA and RNA targeting, both in vivo and in vitro. Base modifications can be used for the tuning of base pairing. For example the targeting of RNA in vivo can be achieved using antisense and/or siRNA oligonucleotides. To enhance hybridization affinity, BNAs can be incorporated at strategic locations with the oligonucleotide sequence of oligonucleotide probes.
Table 1: Tools for targeting DNA and RNA
|
|
Natural
|
Artificial
|
|
Nuclease resistance
|
Poor
(Natural phosphoester linkage)
|
Good
(non-natural backbone)
|
|
Sequence recognition
|
Fixed
(Four bases)
|
Tunable
(modified base = BNA)
|
|
Recognition of mismatched base pair
|
Normal
|
Good
(flexibility of backbone)
|
|
Toxicity
|
Low
|
Usually low, but depends on the modification used.
|
Table 2: BNANC [Me] as tools for targeting DNA and RNA
|
|
Natural
|
N-methyl-BNA
(BNANC[Me])
|
|
RNA selectivity
|
Normal
|
Very Good
|
|
Nuclease resistance
|
Poor
(Natural phosphoester linkage)
|
Very Good
(non-natural backbone)
|
|
Binding Affinity
|
Normal
|
Strong
|
|
Reduction of off-target effects
|
Normal
|
Good
|
|
Transfection
|
Good
|
Very Good
|
|
Compatibility with enzymes
|
Good
|
Good
|
|
Probes
|
Very suitable
|
Very suitable
|
|
Antisense Oligos
|
Usable
|
Very suitable
|
|
siRNA oligos
|
Usable
|
Very suitable
|
|
Sequence recognition
|
Fixed
(Four bases)
|
Tunable
(number of modified bases)
|
|
Recognition of mismatched base pair
|
Normal
|
Good
(flexibility of backbone)
|
|
Toxicity
|
Low
|
Very low
|
Backbone modifications
The bridging of the 2’-oxygen of the ribose with the 4’-carbon in bridged nucleic acids (BNAs) results in a 3’endo (N-type) conformation. This bridging locks the ribose sugar into the N-type conformation. When the bridge contains a methylene linkage between the 2’-oxygen and the 4’-carbon on the ribose the bridged nucleic acids is known as LNA.
Sugar rings are building blocks of oligonucleotides located between the nucleic acid nucleobase and phosphate backbone where they act as flexible links. Ribose and 2’-deoxyribose are the basic subunits that differentiate RNA and DNA. Ribose sugar rings can adopt different conformations and thereby influence the global structure of nucleic acids. The five-membered sugar ring in oligonucleotides is inherently nonpolar. The ribose ring structure can interconvert into different conformations by “pseudorotation.” The conformation with the lowest energy is usually a ring structure with one atom out of plane and four atoms in plane. The process of pseudorotation results in a ring pucker motion. Nonbonding interactions between substituents at the four ring carbon atoms are the cause for puckering resulting in nonplanar ring structures. The pseudorotation cycle describes the interconversion into different puckering modes as they have been observed in nucleic acid structures (see Saenger in Principles of Nucleic Acid Structure, page 57).
The assumption based on structural analysis is that the ribose ring exists in a two-state conformational equilibrium between one N-type and one S-type conformation. If the ribose ring is fused to a second ring the conformation can be fixed.
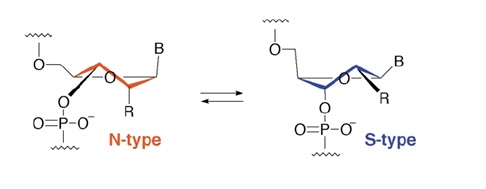
Figure 1: Ribose sugar puckering modes in RNA and DNA. N- and S-type sugar puckering. The N-type conformation (C3-endo or A-form) exists predominantly in A-RNA and the S-type conformation (C2-endo or B-form) in duplexes with B-DNA helical structure.
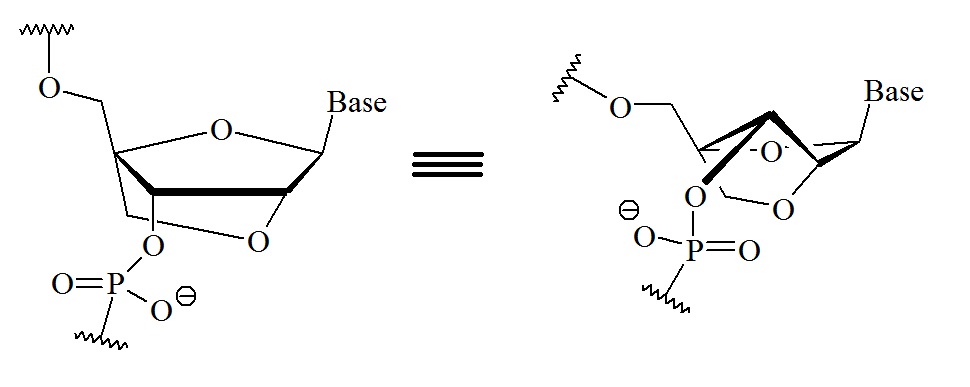
Figure 2: Chemical structure for LNA. A: Typical structure used in most reviews. B: Structure illustrating the N-type conformation.
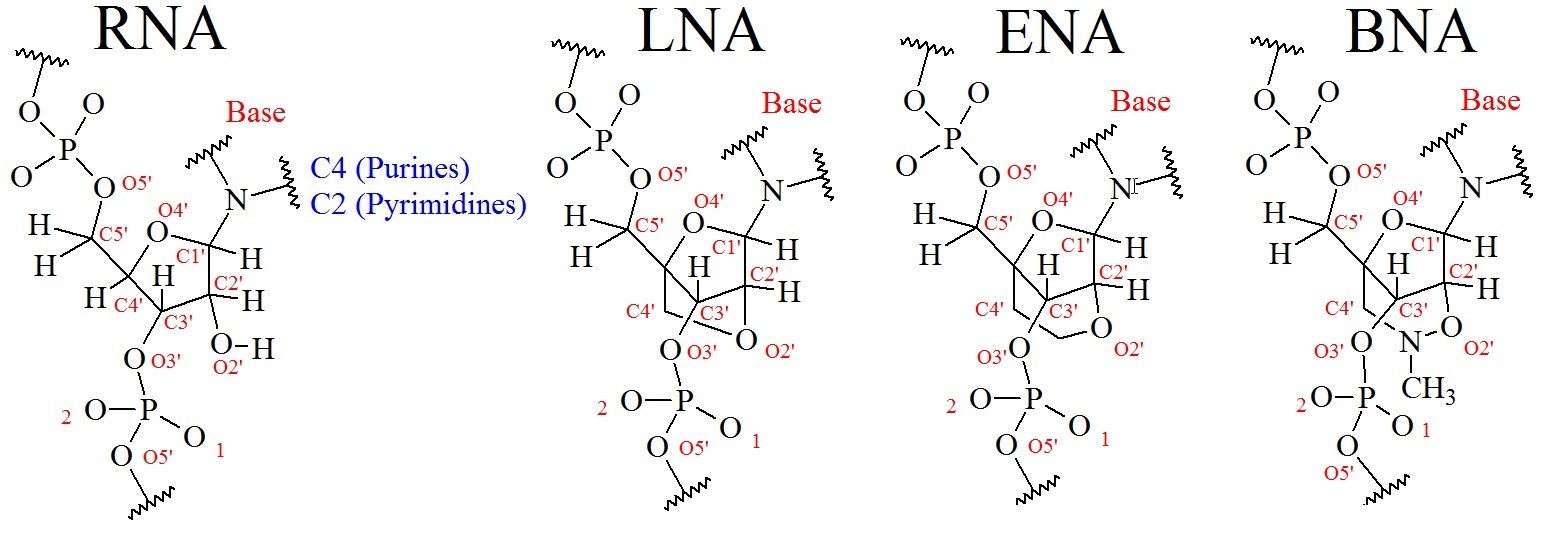
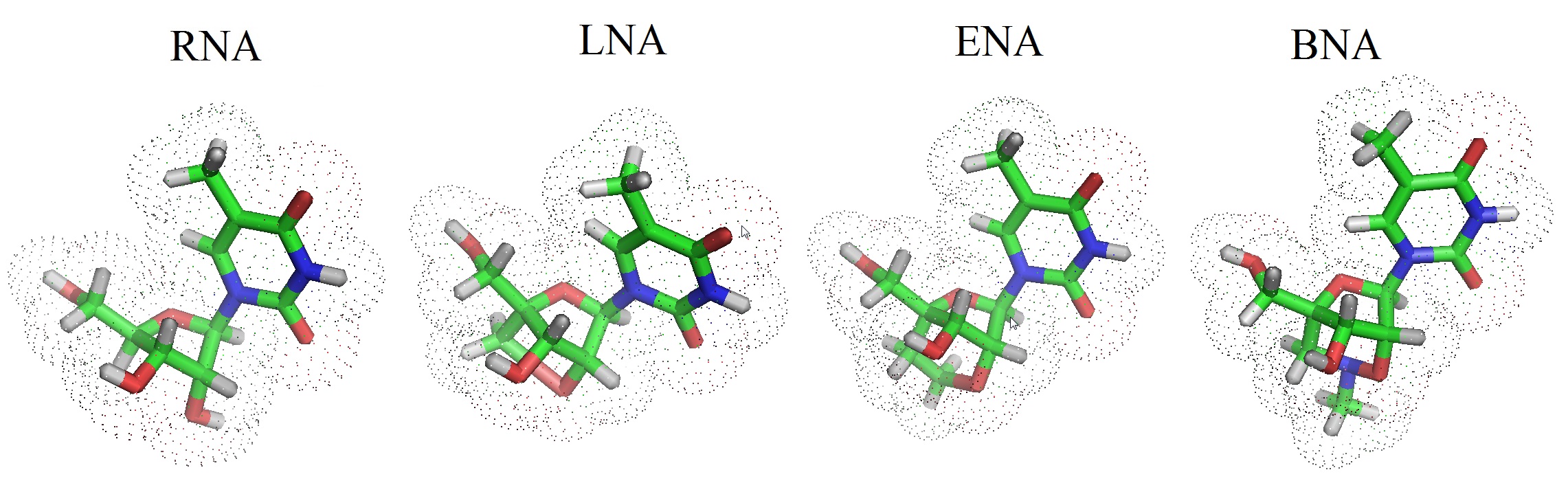
Figure 3: Chemical structures for RNA and various bridged nucleic acids. The numbering nomenclator is shown in the upper panel and the models are shown in the lower panel. The stick models were created using Pymol in the builder and sculpting mode. RNA = ribonucleic acids; LNA = locked nucleic acid; ENA = ethylene nucleic acid; BNA = bridged nucleic acid.
BNA Applications
· Duplex Formation through Hybridization;
· RNA targeting;
· Gene silencing;
· Antisense;
· siRNA;
· Aptamer capping;
· Probes for real-time qPCR;
· BNAClampTM PCR;
· Probes for in-situ hybridization (ISH or FISH)
· Triplex-Forming Oligonucleotides (TFO), thermodynamically favored;
· RNase H Activation;
· Enhanced Nuclease Resistance and Serum Stability;
· Cells Delivery;
· Low In Vivo Toxicity;
· Development of Therapeutic and Antisense Agents;
· BNAzymes.
· Tools for Antigene Strategies, targeting of long noncoding RNAs (lncRNAs);
· Design of BNA aptamers;
· Design of novel reagents and probes for diagnostics;
· Single Nucleotide (SNP) Polymorphism (SNP) Detection;
· RNA Capture Probes, miRNA Detection.
|
Therapeutic Applications
|
|
|
|
|
Antisense
|
Antigene
|
|
|
|
|
Antisense Oligonucleotides
|
Triplex Forming Oligonucleotides (TFO)
|
|
siBNAs
|
Aptamer based approaches
|
|
BNAzyme
|
Transcription Elongation Inhibitors
|
|
Targeting non-coding RNAs
|
Strand Invasion
|
|
Gapmers
|
|
Reference
Ming Huang, Timothy J. Giese, Tai-Sung Lee, and Darrin M. York; Improvement of DNA and RNA Sugar Pucker Profiles from Semiempirical Quantum Methods. J Chem Theory Comput. 2014 Apr 8; 10(4): 1538–1545. Published online 2014 Mar 3. doi: 10.1021/ct401013s PMCID: PMC3985690. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3985690/]
Mayer, G.; The Chemical Biology of Nucleic Acids. 2010. WILEY. ISBN: 978-0-470-51974-5.
---...---