Invader probes are oligonucleotides or modified oligonucleotides used in invader assays for genotyping of single point mutations (SNPs) and gene expression monitoring. Some invader probes also utilize intercalation, the insertion of a molecule between the planar bases of DNA, to recognize specific regions in DNA, such as chromosomal DNA.
One Invader assay uses a structure-specific flap endonuclease (FEN) to cleave a three-dimensional complex formed by hybridizing allele-specific overlapping oligonucleotides to target DNA containing a single nucleotide polymorphism (SNP) site. The basic assay utilizes two synthetic oligonucleotides, one invasive and a signal oligonucleotide probe. Both probes anneal in tandem to the target strand to form the overlapping complex.
The signal probe contains two sequence regions,
(i) a target-specific regioan complementary to the target sequence, and
(ii) a 5'-arm or flap that is noncomplementary to both the target and the invasiv probe sequence (Figure 1).
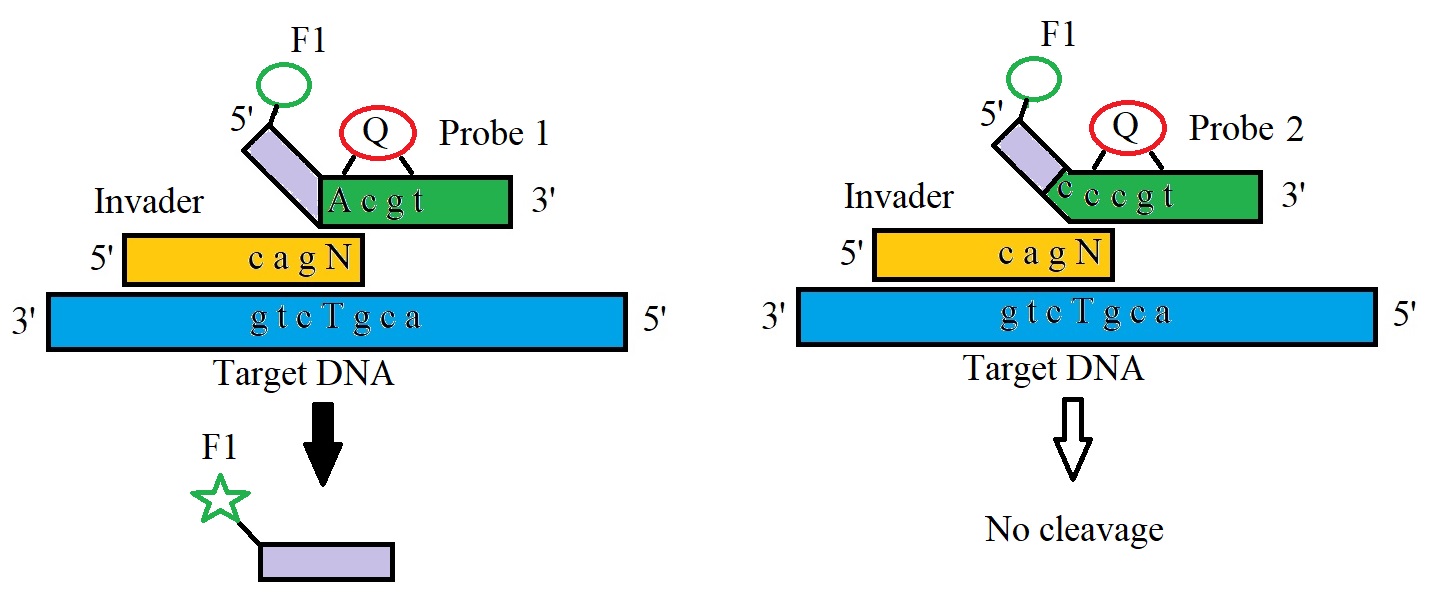
Figure 1: Allele-specific cleavage in an Invader reaction by flap endonucleases (FENs) (Adapted from Oliver 2005).
The annealing of the oligonucleotide complementary to the SNP allele in the target molecule triggers the cleavage of the oligonucleotide by cleavase, a thermostable FEN (Flap endonuclease) that recognizes the overlapping complex (Figure 2).
In 2003, Lyamichev & Neri published a protocol in Method in Molecluar Biology descibing the basic Invader assay.The assay utilizes two oligonucleotide probes that hybridize to target DNA containing a polymorphic site. The two oligonucleotides hybridize to the single-stranded target and form an overlapping invader structure at the site of the SNP. The cleavage of the oligonucleotide flap is detected by (i) a fluorescence resonance energy transfer (FRET) cassette to generate a fluorescent signal or (ii) with the help of fluorescence polarization (FP) probes or by (iii) mass spectrometry. Figure 1 illustrates the allele-specific cleavage in an invader reaction by flap endonucleases.
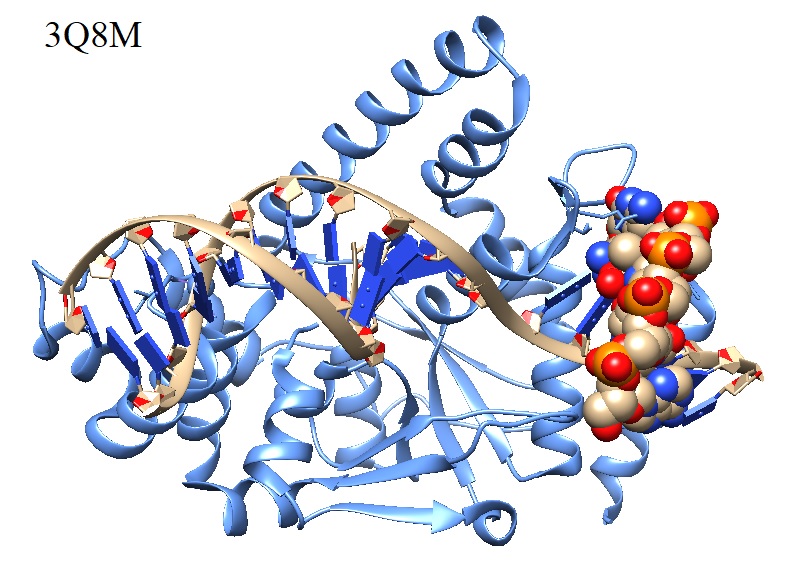
The structure of human flap endonuclease Fen 1 in complex with substrate Flap DNA is shown in figure 2.
|
|
Figure 2: Human flap endonuclease Fen 1 (D181a) in complex with substrate 5’-Flap DNA and K+.
Flap endonuclease (FEN1) is essential for DNA replication and repair by removing RNA and DNA 5' flaps. FEN1 5' nuclease superfamily members acting in nucleotide excision repair (XPG), mismatch repair (EXO1), and homologous recombination (GEN1) incise structurally distinct bubbles, ends, or Holliday junctions.
(Tsutakawa et al. 2011. PDB ID 3Q8M)
|
Reference
Germer JJ, Majewski DW, Yung B, Mitchell PS, Yao JD. Evaluation of the invader assay for genotyping hepatitis C virus. J Clin Microbiol. 2006 Feb;44(2):318-23. [PMC]
Hsu TM, Law SM, Duan S, Neri BP, Kwok PY. Genotyping single-nucleotide polymorphisms by the invader assay with dual-color fluorescence polarization detection. Clin Chem. 2001 Aug;47(8):1373-7. [PubMed]
Lyamichev, V., Neri, B. (2003). Invader Assay for SNP Genotyping. In: Kwok, PY. (eds) Single Nucleotide Polymorphisms. Methods in Molecular Biology™, vol 212. Springer, Totowa, NJ. [MMB 212].
Michael Olivier; The Invader® assay for SNP genotyping.Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. Volume 573, Issues 1–2, 3 June 2005, Pages 103-110. [PMC]
Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, Cooper PK, Grasby JA, Tainer JA. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011 Apr 15;145(2):198-211. [PubMed]
---...---
Bio-Synthesis provides a full spectrum of oligonucleotide and peptide synthesis including bio-conjugation services as well as high quality custom oligonucleotide modification services, back-bone modifications, conjugation to fatty acids and lipids, cholesterol, tocopherol, peptides as well as biotinylation by direct solid-phase chemical synthesis or enzyme-assisted approaches to obtain artificially modified oligonucleotides, such as BNA antisense oligonucleotides, mRNAs or siRNAs, containing a natural or modified backbone, as well as base, sugar and internucleotide linkages.
The synthesis of FRET-oligonucleotides or peptides is also possible.
Bio-Synthesis also provides biotinylated mRNA and long circular oligonucleotides.
---...---