In humans, iron is a vital trace element. Iron is essential in oxygen transport, oxidative metabolism, cellular proliferation, as well as for many catalytic reactions. Iron metabolism appears to be a complicated process and needs to be tightly controlled and maintained within the ideal range for optimal iron homeostasis. Since no active mechanism for iron excretion exists, the absorbed amount of iron by the intestine is tightly controlled (Yiannikourides, Latunde-Dada; 2019).
Genes critical for the uptake, sequestering, and utilization of iron are post-transcriptionally regulated. The mRNAs of these genes contain conserved stem-loop structures called iron-responsive elements (IREs). Iron-responsive elements are found in untranslated regions of mRNAs of proteins involved in iron metabolism. IREs are nucleic-acid binding sites for a cytosolic RNA-binding protein called the IRE-binding protein (IRE-BP). The iron status of a cell reversibly regulates the binding of IRE-BP to IREs. The iron-responsive element (IRE) is found in the 5’-untranslated region (5’-UTR) of ferritin messenger RNA (mRNA), delta aminolevulinic acid synthase mRNAs, and in the 3'-UTR of transferrin receptor mRNA.
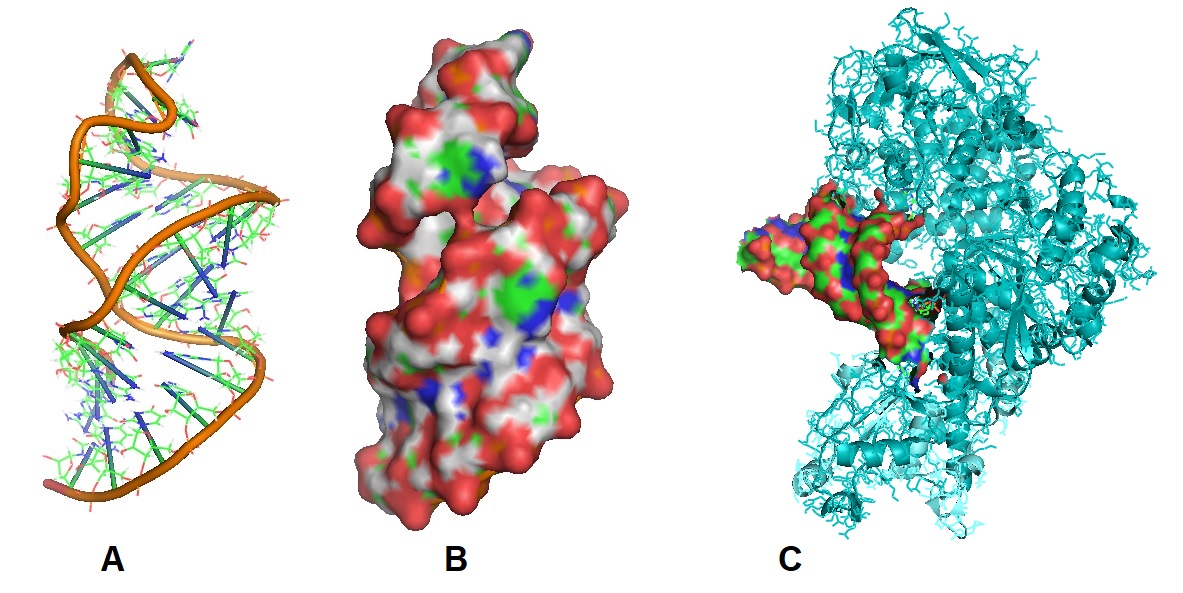
Figure 1: (A and B) Structural models of the Iron Responsive Element RNA (IRE), 1AQO. Sequence: GGA GTG CTT CAA CAG TGC TTG GAC GCT CC. The IRE is an approximately 30 nucleotide RNA hairpin located in the 5’-untranslated (5’-UTR) region of all ferritin mRNAs and in 3’-UTR region of all transferrin receptor mRNAs. IREs are bound by two related IRE-binding proteins (IRPs) which help control intracellular levels of iron by regulating the expression of both ferritin and transferrin receptor genes (Address et al. 1997). (C) Structural model of Iron Regulatory Protein 1 in complex with ferritin IRE-RNA, 3SNP. Iron regulatory protein 1 (IRP1) binds iron-responsive elements (IREs) in messenger RNAs (mRNAs), to repress translation or degradation, or binds an iron-sulfur cluster, to become a cytosolic aconitase enzyme (Walden et al, 2006). Images were created using PyMOL and PDB files.
A part of the 3' untranslated region of the mRNA regulates transferrin receptor mRNA levels post-transcriptional in response to the presence or absence of iron. In-vitro assays showed that IREs from the ferritin and transferrin receptor mRNAs compete for interaction with a cytoplasmic protein, indicating that the activity of the IRE-binding protein dependents on the cells iron status. At low iron concentrations or if the iron is absent, IRE-BPs bind the IRE motif in the ferritin mRNA, causing reduced translation rates. The presence of iron releases the mRNA via interaction with the IRE. Furthermore, the transferrin receptor mRNA is more stable when IRE-BPs are bound to the IREs 3'-UTR of transferrin receptor mRNA.
RNA sequences with stem-loop structures, as found in iron-responsive elements look like naturally occurring aptamers. Regulatory RNA sequence motifs are similar to aptamers made of single-stranded oligonucleotides with sequences that precisely fold into a well-defined three-dimensional (3D) structure binding to selected molecules.
Purification of IREs-binding Proteins
Biotinylation of regulatory sequences such as the IREs allows the purification of proteins binding to the regulatory elements. First, prepare biotinylated oligonucleotides that contain the IRE sequence motifs. Select the cells or tissue to be studied. Next, incubate the unfractionated cytosol with the biotinylated oligonucleotides with a solution of avidin. Capture specific RNA-protein complexes with the help of biotin-agarose beads. Release the proteins from the RNA in the presence of high salt. The second round of purification will yield relative homogeneous protein solutions.
Rouault et al. used this approach for the purification and identification of the protein in the human liver that binds to the iron-responsive RNA sequences. Furthermore, gel-mobility shift assays or cross-linking induced by UV irradiation allow the study of RNA-protein interaction.
Biotinylated-oligonucleotides containing regulatory elements stabilized at selected nucleotide positions with bridged-nucleic acids (BNAs) may allow enhanced purification of regulatory RNA-binding proteins.
Reference
Addess KJ, Basilion JP, Klausner RD, Rouault TA, Pardi A; Structure and dynamics of the iron responsive element RNA: implications for binding of the RNA by iron regulatory binding proteins. J Mol Biol. 1997 Nov 21;274(1):72-83. [Pubmed]
John L. Beard, Iron Biology in Immune Function, Muscle Metabolism and Neuronal Functioning, The Journal of Nutrition, Volume 131, Issue 2, February 2001, Pages 568S–580S. [Pubmed]
Casey JL, Koeller DM, Ramin VC, Klausner RD, Harford JB. Iron regulation of transferrin receptor mRNA levels requires iron-responsive elements and a rapid turnover determinant in the 3' untranslated region of the mRNA. EMBO J. 1989 Dec 1;8(12):3693-9. PMID: 2583116; PMCID: PMC402052. [Pubmed]
Harrell CM, McKenzie AR, Patino MM, Walden WE, Theil EC. Ferritin mRNA: interactions of iron regulatory element with translational regulator protein P-90 and the effect on base-paired flanking regions. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4166-70. doi: 10.1073/pnas.88.10.4166. PMID: 1903535; PMCID: PMC51619. [Pubmed]
Rouault TA, Hentze MW, Haile DJ, Harford JB, Klausner RD. The iron-responsive element binding protein: a method for the affinity purification of a regulatory RNA-binding protein. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5768-72. doi: 10.1073/pnas.86.15.5768. PMID: 2474819; PMCID: PMC297711. [Pubmed]
Yiannikourides, A.; Latunde-Dada, G.O. A Short Review of Iron Metabolism and Pathophysiology of Iron Disorders. Medicines 2019, 6, 85. [PMC]
---...---