Using mass spectrometry in combination with X-ray crystallography, Wang et al. recently reported a method called CapQuant that allows the identification of cap-like RNAs in bacteria, virusus, yeast and human tissues. Bacterial RNA polymerase (RNA Pol) can initiate transcription in vitro by accepting nucleotide metabolites capped with flavin adenine dinucleotide (FAD), uridine diphosphate glucose (UDP-Glc), and uridine diphosphate N-acetylglucosamine (UDP-GlcNAc).
Messenger RNA (mRNA) regulates cell proliferation. The 5’- terminal ends of cellular mRNAs contain an m7GpppN cap, in which N can be any nucleotide. The RNA helicase eIF4A and the scaffold protein eukaryotic translation initiation factor 4G (eIF4G) and the capping protein eIF4E are part of the complex that loads the mRNAs onto the 40 S ribosomal subunit, together with eIF3. eIF4E has a crucial role in the regulation of translation.
4E-binding proteins (4E-BPs) block the interaction of eIF4E with eIF4G to negatively regulate the formation of the eIF4F complex. The "closed-loop" model explains that the induction of 5'- to 3'-proximity of mRNAs requires the initiation factors eIF4E, eIF4G, and PABP. The model proposes the cap's interactions, with eIF4E, eIF4G, PABP, and the poly(A) tail and that the disruption of these interactions leads to functional effects. PABP binding to poly(A) increases PABP's affinity to eIF4G and the affinity of eIF4E for the cap as well.
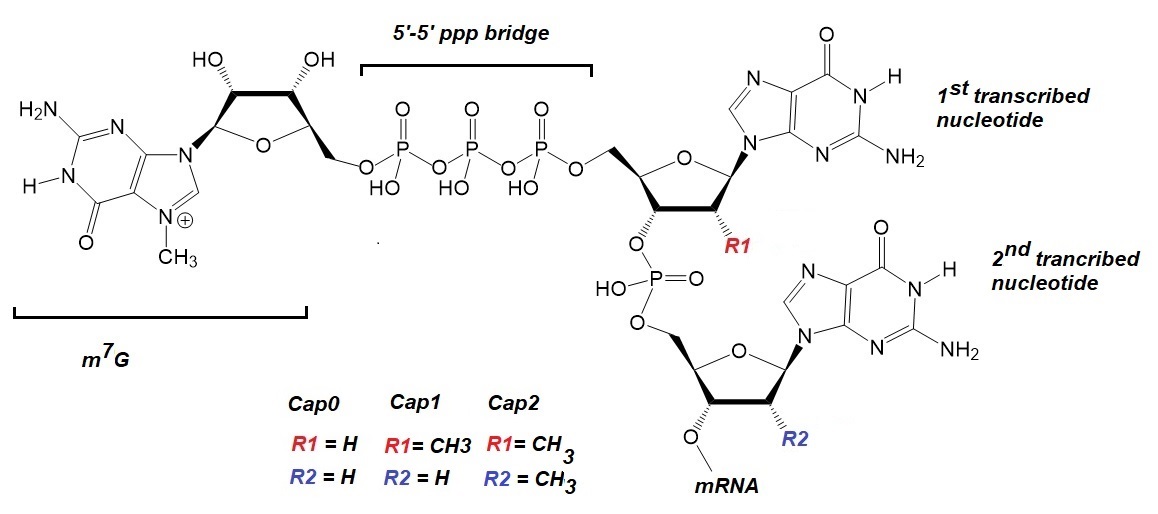
Figure 1: Chemical structures of mRNA Cap0, Cap1, and Cap2.
Figure 2: Brown et al. studied the interaction of synthetic N7-substituted GMP cap anlogs with the eIF4E mononucleotide binding site..
RNA-based pharmaceutical therapeutics and vaccines are a new approach to treating chronic and rare diseases, including COVID-19. Fore example, the deregulation of translation control appears the cause of many cancer types. The expression of eIF4E influences cell growth and phenotype. The overexpression of eIF4E leads to accelerated cell division and malignant transformation.
Structural complexes between proteins and their substrates can be studied in the gas-phase to determine equilibrium dissociation constants. Brown et al., in 2007, showed that mass spectrometry allows the measurement of the apparent gas-phase equilibrium dissociation constants (Kd) values for the specific molecular binding events. The Kds of guanosine triphosphate (GTP), GMP, and cap derivatives interactions with eIF4E were determined. Figure 3 shows molecular models of the cystal structure of eIF4E, in complex with the cap m7GpppA, and a 4EBP1 peptide. Figure 4 illustrates the interactions between eIF4E, eIF4G, and the cap analog m7GTP as well as the interactions between the eIF4E dimer, eIF4G, and the cap analog (See Peter at al. for more detail).
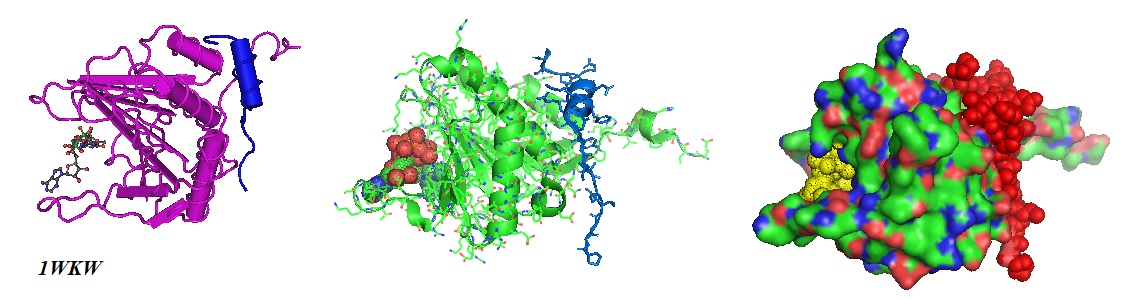
Figure 3: Molecular models from the crystal structure of the ternary complex of eIF4E, m7GpppA and 4EBP1 peptide PGGTRIIYDRKFLMECRNSP (blue and red) (PDB ID 1WKW). The cap m7GpppA (yellow) is shown in the binding pocket of the protein (Tomoo et al. 2005; PDB ID 1WKW).
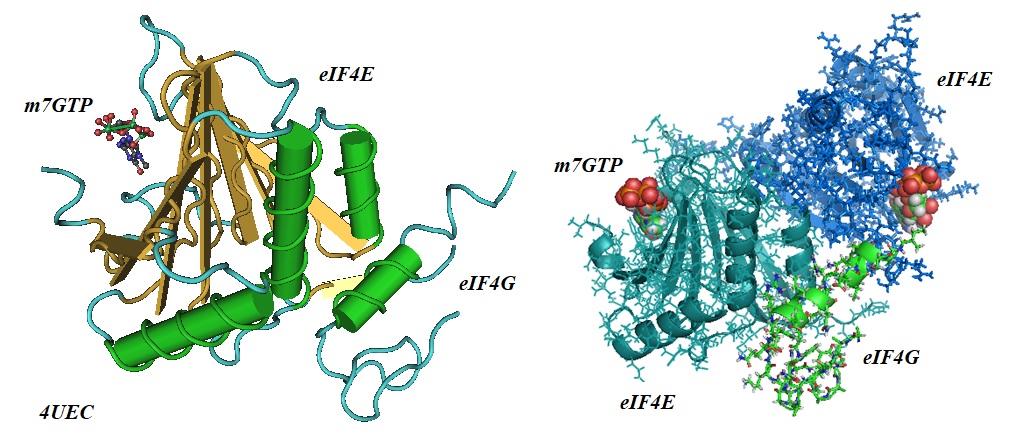
Figure 4 : Eukaryotic Translation Initiation Factor 4E (eIF4E; magenta) in complex with an isoform of eukaryotic translation initiation factor 4G (eIF4G; blue), and cap analog m7GTP (PDB ID 4EUC). Left: The interactions between eIF4E, eIF4G, and the cap analog m7GTP is ilustrated. Right: The interactions between the eIF4E dimer, eIF4G, and the cap analog m7GTP is illustrated (See Peter at al. for more detail).
Reference
Brown CJ, McNae I, Fischer PM, Walkinshaw MD; Crystallographic and mass spectrometric characterisation of eIF4E with N7-alkylated cap derivatives. J Mol Biol (2007) 372 p.7-15. [PubMed]
Daniel Peter, Cátia Igreja, Ramona Weber, Lara Wohlbold, Catrin Weiler, Linda Ebertsch, Oliver Weichenrieder, Elisa Izaurralde; Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol Cell. 2015 Mar 19;57(6):1074-1087. [PubMed]
Koji Tomoo Yasunori Matsushita, Hiroyuki Fujisaki, Fumi Abiko, Xu Shen, Taizo Taniguchi, Hiroo Miyagawa, Kunihiro Kitamura, Kin-ichiro Miura, Toshimasa Ishida; Structural basis for mRNA Cap-Binding regulation of eukaryotic initiation factor 4E by 4E-binding protein, studied by spectroscopic, X-ray crystal structural, and molecular dynamics simulation methods. Biochim Biophys Acta. 2005 Dec 1;1753(2):191-208. [PubMed]
Jin Wang, Bing Liang Alvin Chew, Yong Lai, Hongping Dong, Luang Xu, Seetharamsingh Balamkundu, Weiling Maggie Cai, Liang Cui, Chuan Fa Liu, Xin-Yuan Fu, Zhenguo Lin, Pei-Yong Shi, Timothy K. Lu, Dahai Luo, Samie R. Jaffrey, Peter C. Dedon; Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. bioRxiv 683045. [Nucleic Acids Research]
---...---