Utilizing UV absorbance spectrophotometry allows for the rapid quantification of nuclei acids. This type of analysis is the most popular analytical method used. UV absorbance spectrophotometry enables quantifying nucleic acids, such as DNA and RNA. The process allows quantifying nucleic acids based on their ability to absorb UV light, which is directly proportional to their concentration.
Nucleic acids absorb UV light because they contain aromatic bases in their structure. Adenine, guanine, cytosine, and thymine (in DNA) or uracil (in RNA) have a characteristic absorption spectrum in the UV region, with a maximum absorbance at around 260 nm.
The amount of UV light absorbed at this wavelength is directly proportional to the concentration of nucleic acids in the sample and the light path length. The linearity of the measured absorbance value allows for calculating nucleic acid concentration in the sample.
However, sample impurities such as proteins and other organic molecules can interfere with the measurement.
Method 1 (Cavaluzzi & Borer, 2004):
This method ignores base composition and assumes that the average molar mass and extinction coefficient of nucleotides is 330 g/mol and 10 mmol-1cm-1, respectively.
For an A260 nm absorbance of 1, a concentration of 33 μg/ml is obtained for a single-stranded oligonucleotide using the Lambert-Beer equation.
Method 2 (Kallansrud & Ward 1996):
This method assumes that ɛ is the sum of nucleotide extinction coefficients weighted by the number of times each base appears in the sequence.
Method 1 and 2 do not account for potential hypochromicity in the measured oligonucleotide or complex nucleic acid structures. However, for accurate quantification, the hypochromicity of the oligonucleotides needs to be considered.
Method 3:
This method uses a near-neighbor calculation to account for hypochromicity. Based on published data, this approach yields extinction coefficients within 20% of experimentally measured extinction coefficients.
Hypochromic measurements are made by comparing the absorbance of non-denatured and denatured nucleic acids and determining the melting profile using UV spectrophotometry. Unfortunately, at high temperatures, partial hydrolysis of RNA can occur. At moderate temperatures, complete denaturation is not guaranteed. Also, large dsRNA requires high temperatures for denaturation, and the extinction coefficients are affected by temperature.
Method 4 (Wilson et al., 2014):
Determination of RNA concentration by thermal hydrolysis.
Using the absorbance values for hydrolyzed oligonucleotides allows for a more accurate calculation of concentrations present in a sample.
For this method, measure the UV absorbance at 260 nm with a Nanodrop microvolume UV/VIS spectrometer.
[1] Start with an aliquot of RNA sample (~2 μl).
[2] Dilute the sample to a starting A260 of ~10 AU in hydrolysis buffer. Mix equal amounts of sample solution (2 μl) with 500 mM Na2CO3, and 100 mM EDTA at pH 7-9 (2μl) and add sterile water (16 μl) in a reaction vial with a safety lock lid (Total volume 20 μl).
[3] Incubate at 95ºC for 90 minutes in a dry-heat block.
[4] Cool the reaction mixture and briefly centrifuge prior to opening.
[5] Neutralize the hydrolysis solution using 0.1 M acetic acid if the hydrolysis was performed at a higher pH (pH 8 to 9).
[6] Measure UV absorbance at 260 nm using 2 μl aliquots. To minimize errors, measure three different aliquots.
[7] Calculate the concentration of the RNA sample.
[8] Calculations:
The concentration of RNA sample = A260 value for hydrolyzed divided by the product of the sum of the number of nucleotides present in the sequence and published extinction coefficients for the 5’-mononucleotides.
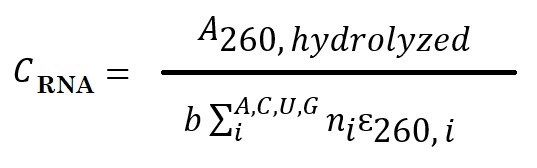 ,
,
Where b is the path length, i is the nucleotide identity (A, C, U, or G), ni is the number of nucleotides i present in the sequence, and ɛi is the extinction coefficient for the given mononucleotide found in the literature. Use the following extinction coefficients for the 5′ mononucleotides to approximate the extinction coefficients for the hydrolysis products: pA = 15,020 M−1cm−1, pC = 7,070 M−1cm−1, pG = 12,080 M−1cm−1, pU = 9,660 M−1cm−1 (Cavaluzzi & Borer, 2004).
Method 5 (Nwokeoji et al., 2023):
This method utilized the chemical denaturant dimethyl sulfoxide (DMSO) and a short thermal denaturation step. DMSO prevents renaturation of the duplex nucleic acids (dsDNA/RNA). The absorbance of unstructured and structured nucleic acids is accurately measured to determine their hypochromicity and extinction coefficients.
Using this method, Nwokeoji et al. determined extinction coefficient values of 46.18 - 47.29 μg/mL/A260 for dsRNA.
References
Adler AJ, Greenfield NJ, Fasman GD. Circular dichroism and optical rotatory dispersion of proteins and polypeptides. Methods Enzymol. 1973;27:675-735. [sciencedirect]
Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5'-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004 Jan 13;32(1):e13. [PMC]
Handbook of Biochemistry and Molecular Biology (R. Lundblad and F. Macdonald (eds.), 4th Edn., CRC Press, Taylor & Francis Group, Boca Raton-London-New York, 2010.
Kallansrud G, Ward B. A comparison of measured and calculated single- and double-stranded oligodeoxynucleotide extinction coefficients. Anal Biochem. 1996 Apr 5;236(1):134-8.
Lee J, Vogt CE, McBrairty M, Al-Hashimi HM. Influence of dimethylsulfoxide on RNA structure and ligand binding. Anal Chem. 2013 Oct 15;85(20):9692-8. [PMC]
Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13(6):515-37. [liebertpub]
Nwokeoji AO, Kilby PM, Portwood DE, Dickman MJ. Accurate Quantification of Nucleic Acids Using Hypochromicity Measurements in Conjunction with UV Spectrophotometry. Anal Chem. 2017 Dec 19;89(24):13567-13574. [ACS]
Strauss, J. H., Jr., Kelly, R. B., Sinsheimer, R. L.: Denaturation of RNA with dimethyl sulfoxide. Biopolymers 6, 793–807 (1968). [onlinelibrary]
Wilson SC, Cohen DT, Wang XC, Hammond MC. A neutral pH thermal hydrolysis method for quantification of structured RNAs. RNA. 2014 Jul;20(7):1153-60. [PMC]
---...---