Peptide linkers or drug-peptide linkers enable the development of next-generation antibody-drug conjugates (ADCs) with enhanced plasma stability and function. In addition, linker peptides also allow the design and development of fusion proteins, protein conjugates, and oligonucleotide conjugates, enabling new therapeutics development. A thorough understanding of peptide linker structures, conformations, and functions will significantly facilitate the construction of stable and bioactive recombinant fusion proteins and similar biomolecules for drug delivery applications.
Flexible peptides can link proteins, protein domains, cyclic peptides, oligonucleotides, or proteins and oligonucleotides. Peptide linker sequences that naturally occur in protein complexes stabilize protein structures to allow these to function optimally.
Bifunctional fusion proteins:
1. Extend plasma half-life by decreasing access to proteases.
2. Decrease renal filtration.
3. Alter intracellular routing via receptor-mediated recycling enables absorption across epithelial bilayers by binding
to transcytosis receptors.
4. Allow targeting over-expressing in vivo sites or uniquely expressing specific receptors or antigens.
Recent advancements in biotechnology allow the design and synthesis of fusion proteins for novel protein therapeutics that can improve the performance of current protein drugs.
Natural occurring peptide linkers
Natural linker peptides adopt a variety of conformations in secondary structure. Functional proteins contain these peptides sequences as helices, β-strands, coils, bends, and turns. The average length of peptide linkers in natural multi-domain proteins is approximately 6.5 residues according to Argos, P. and 10.0 ± 5.8 residues according to George and Heringa. George and Heringa grouped most natural linkers into helical and non-helical. The α-helical conformation is a rigid and stable structure containing intra-segment hydrogen bonds and a closely packed backbone. Linkers with α-helices can serve as rigid spacers reducing protein domain interactions. Non-helical linkers tend to be rich in prolines. Prolines tend to increase the stiffness of the linker moiety. However, the peptide linker's length, composition, hydrophobicity, and secondary structure are critical for proteins to function correctly (Argos, P. 1990; George & Heringa, G.R. 2022).
The average hydrophobicity of peptide linkers decreases with the increase in length. Longer linkers are more hydrophilic and more exposed to the aqueous solvent than shorter linkers. Natural multi-domain proteins contain proline-rich sequences as interdomain linkers. One example is the linker between the lipoyl and E3 binding domain in pyruvate dehydrogenase, GAAPAAAPAKQEAAAPAPAAKAEAPAAAPAAKA. Another example is the proline 9 (PPPPPPPPP) linker between the central and the C-terminal domains in cysteine proteinase.
The proline P9 linker is also found in the human butyrylcholinesterase tetramer. The C-terminal tryptophan amphiphilic tetramerization (WAT) helices from each subunit mediate tetramerization to assemble the complex. The protein complex is assembled around a central lamellipodia-derived oligopeptide with a proline 12 peptide attachment domain (PRAD) sequence. The proline peptide adopts a polyproline II helical conformation and runs antiparallel. A homologous proline linker is present in the Transformation/transcription domain-associated protein of the human SAGA coactivator complex (TRRAP, core, splicing module) [PDB ID 7KTS] and in Leiomodin-2 in the actin-leiomodin-2 complex [PDB ID 4RWT]. However, the linker peptide is flexible in all structures and, therefore, not observed in the solved x-ray structures.
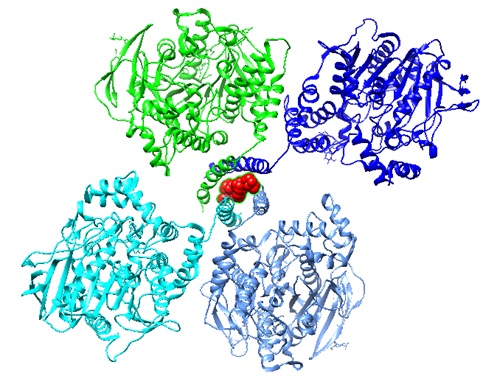
Figure 1: Structure of the fully glycosylated human butyrylcholinesterase tetramer (PDB ID 6I2T; Leung et al. 2018). The proline peptide is shown in red.
Linkers containing small, polar amino acids such as threonine or serine and glycine are favored because they provide good flexibility due to their small sizes. Furthermore, these linker types may help maintain the stability of the linker structure in aqueous solvents through the formation of hydrogen bonds with water (Whitley et al. 2018; Mottram JC et al. 1989).
As an essential part of recombinant fusion proteins, linkers allow the construction of stable, bioactive fusion proteins. The lipoyl, subunit-binding and catalytic domains of the dihydrolipoamide acetyltransferase subunits (E2p) of the Escherichia coli pyruvate dehydrogenase complex are connected by linker peptide sequences. These linkers are characteristically rich in alanine and proline residues (Turner et al. 1993; Chen et al. 2013). Table 1 shows a list of flexible, rigid, and cleavable linkers utilized in fusion proteins.
Table 1: A list of linkers and their functionalities (Adapted from Chen et al. 2013 [PMC]).
|
Linker
Function
|
Fusion Protein
|
Linker
|
|
Type
|
Sequencea
|
|
Increase
Stability/Folding
|
scFv
|
flexible
|
(GGGGS)3
|
|
G-CSF-Tf
|
flexible
|
(GGGGS)3
|
|
HBsAg preS1
|
flexible
|
(GGGGS)3
|
|
Myc- Est2p
|
flexible
|
(Gly)8
|
|
albumin-ANF
|
flexible
|
(Gly)6
|
|
virus coat protein
|
rigid
|
(EAAAK)3
|
|
beta-glucanase-xylanase
|
rigid
|
(EAAAK)n (n=1-3)
|
|
Increase
expression
|
hGH-Tf and Tf-hGH
|
rigid
|
A(EAAAK)4ALEA(EAAAK)4A
|
|
G-CSF-Tf and
Tf-G-CSF
|
rigid
|
A(EAAAK)4ALEA(EAAAK)4A
|
|
Improve
biological
activity
|
G-CSF-Tf
|
flexible
|
(GGGGS)3
|
|
G-CSF-Tf
|
rigid
|
A(EAAAK)4ALEA(EAAAK)4A
|
|
hGH-Tf
|
rigid
|
A(EAAAK)4ALEA(EAAAK)4A
|
|
HSA-IFN-α2b
|
flexible
|
GGGGS
|
|
HSA-IFN-α2b
|
rigid
|
PAPAP
|
|
HSA-IFN-α2b
|
rigid
|
AEAAAKEAAAKA
|
|
PGA-rTHS
|
flexible
|
(GGGGS)n (n=1, 2, 4)
|
|
interferon-γ-gp120
|
rigid
|
(Ala-Pro)n (10 – 34 aa)
|
|
GSF-S-S-Tf
|
cleavable
|
disulfide
|
|
IFN-α2b-HSA
|
cleavable
|
disulfide
|
|
Allow targeting
|
FIX-albumin
|
cleavable
|
VSQTSKLTR↓AETVFPDVb
|
|
LAP-IFN-β
|
cleavable
|
PLG ↓ LWA c
|
|
MazE-MazF
|
cleavable
|
RVL↓AEA; EDVVCC↓SMSY;
GGIEGR↓GSc
|
|
Immunotoxins
|
cleavable
|
TRHRQPR↓GWE;
AGNRVRR↓SVG;
RRRRRRR↓R↓Rd
|
|
Immunotoxin
|
cleavable
|
GFLG↓e
|
|
Alter PK
|
|
dipeptide
|
LE
|
|
G-CSF-Tf and hGH-Tf
|
rigid
|
A(EAAAK)4ALEA(EAAAK)4A
|
| |
cleavable
|
Disulfide
|
aProtease sensitive cleavage sites are indicated with “↓”; bFactor XIa/FVIIa sensitive cleavage; cMatrix metalloprotease-1 sensitive cleavage sequences, one example provided here; dHIV PR (HIV-1 protease); NS3 protease (HCV protease); Factor Xa sensitive cleavage, respectively; eFurin sensitive cleavage; fCathepsin B sensitive cleavage.
Flexible linkers
Flexible linkers allow joining proteins or protein domains that require a certain degree of movement or interaction. Generally, small, non-polar (Gly) or polar (Ser or Thr) amino acids are used. The most used flexible linkers are peptide sequences composed of serine and glycine residues (GS linkers). See table 1. Other types of flexible linkers can also be designed: Adding glycine and serine residues to the linker provide flexibility, and adding glutamic acid and lysine residues improve solubility. The glycine linker (Gly)8 is flexible, increases the accessibility of an epitope to antibodies, and is stable to proteolytic enzymes. As table 1 shows, several glycine-rich linkers enabled the construction of fusion proteins.
Rigid linkers
Rigid linkers keep a fixed distance between protein domains to maintain their independent function. Alpha helix-forming linkers such as (EAAAK)n help construct recombinant fusion proteins. The A(EAAAK)nA (n = 2 to 5) linker has an α-helical conformation stabilized by E--K+ salt bridges. Proline-rich sequences, for example (XP)n, in which X can be any amino acid, but preferable A, K, or E, are also rigid linkers. The (AP)7 dipeptide repeat in the N-terminal alkali light chain of skeletal muscle exhibits an extended and inflexible conformation. The presence of prolines imposes a strong conformational constrain. Rigid linkers have relatively stiff structures by adopting α-helices or containing multiple proline residues. Rigid peptide linkers are critical for the spatial separation of protein domains and the stability or bioactivity of fusion proteins.
In Vivo cleavable linkers
Linker peptides that are not cleavable covalently join biomolecules together, which now act as one molecule throughout the in vivo process. In general, these linkers add a prolonged plasma half-life to the conjugate. Some drawbacks of the non-cleavable linker can be steric hindrance between functional domains, a decreased bioactivity, and altered biodistribution and metabolism of proteins.
Cleavable linkers allow the release of free functional domains in vivo. However, the design of cleavable linkers is challenging. In the case of recombinant fusion proteins, oligonucleotides are used to add linkers between proteins.
Well-designed cleavable linkers take advantage of unique in vivo processes allowing them to be cleaved under specific conditions such as reducing reagents or proteases. A well-studied in vivo process is the reduction of disulfide bonds. A dithiocyclopeptide linker used in fusion proteins optimizes the proteins' biological activity and pharmacokinetic profiles. The disulfide linker LEAGCKNFFPR!SFTSCGSLE is based on a dithiocyclopeptide with an intramolecular disulfide bond formed between two cysteine residues (Chen et al. 2010). This linker also contains a thrombin-sensitive sequence (PRS) between the two cysteine residues. The treatment of fusion proteins containing this linker in vitro with thrombin results in the cleavage of the thrombin-sensitive sequence realizing the two parts of the fusion protein.
The cyclopeptide linker CRRRRRREAEAC contains an intramolecular disulfide bond between two cysteine residues and a peptide sequences sensitive to the secretion of the signal processing proteases in the yeast secretory pathway. During protein expression, the linker is cleaved by protease Kex2 at the arginine upstream of the glutamic acid residue (CRRRRRR!EAEAC), followed by the proteases Kex1 and Ste13. During secretion, the amino acids between two cysteine residues in the liker are removed, and the disulfide-linked fusion protein is expressed in Pichia pastoris (Zhao et al. 2012).
Linker for antibody drug conjugates
The approval of rituximab, a chimeric monoclonal antibody (mAB) that recognizes the CD20 protein in 1997 by the U.S. Food and Drug Administration to treat B-cell non-Hodgkin lymphomas resistant to other chemotherapies ushered in an area in which monoclonal antibodies became important components of therapeutic regimens in oncology. To circumvent obstacles encountered with earlier drugs, showing off-target effects such as the targeting of healthy dividing cells and other severe side effects, researchers turned to antibody-drug conjugates or ADCs to selectively deliver toxic compounds to diseased tissue to use as “Magic Bullets”.
A “Magic Bullet” is a drug or concept first proposed in the early 1900s by Paul Ehrlich, a German physician and scientist, that allows the selective targeting of diseased tissue cells. Antibody-drug-conjugates that exhibit improved potency and effectiveness are now considered to be such magic bullets.
The mAB rituximab is used to treat cancers of the white blood system such as leukemias and lymphomas. Monoclonal antibodies have now become a thriving class of therapeutic molecules. This success resulted from significant progress made in molecular biology and biotechnology, which contributed to our understanding of the inner workings of the human cell and the key players in the immune system.
Therapeutic mABs have substantially affected medical care for a wide range of diseases in the past two decades. The reason is the high specificity and ability of mABs to bind target antigens marking these for removal by methods such as complement-dependent cytotoxicity or antibody-dependent cell-mediated cytotoxicity. In addition, mABs can also have therapeutic benefits by binding and inhibiting the function of target antigens. Unfortunately, as medical scientists have found out, antibodies against tumor-specific antigens often lack therapeutic activities.
In recent years, the conjugation of cytotoxic drugs or radionuclides has expanded the utility of mABS. Antibody-drug-conjugates (ADCs) with improved potency and effectiveness are now used to target and deliver a toxic payload to the selected diseased tissue. In the past decade, antibodies have been conjugated to several cytotoxic drugs using various linker chemistries. Two such drugs are marketed in the United States. These are ado-trastuzumab emtansine (Kadcyla®) and brentuximab vedotin (Adcetris®).
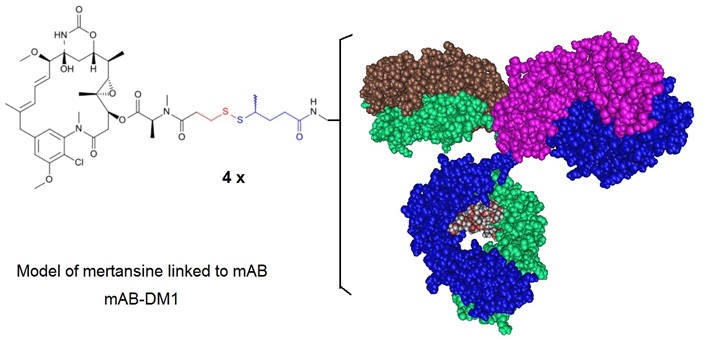
Figure 2: An Antibody Drug Conjugate where a mAb is conjugated to DM1 illustrating the conjugation approach used for Ado-Trastuzumab Emtansine. Ado-trastuzumab emtansine, formerly called Trastuzumab-DM1 (T-DM1), is a HER2 antibody drug conjugate (ADC). In this ADC the trastuzumab antibody is linked to the cell-killing agent, DM1. T-DM1 combines two strategies, (i) anti-HER2 activity and (ii) targeted intracellular delivery of the potent anti-microtubule agent, DM1 (a maytansine derivative) to produce cell cycle arrest and apoptosis. Ado-trastuzumab emtansine is marketed under the brand name Kadcyla and is indicated for use in HER2-positive, metastatic breast cancer patients who have already used taxane and/or trastuzumab for metastatic disease or had their cancer recur within 6 months of adjuvant treatment.
The ADC brentuximag vedotin or Adcetris® combines an anti-CD30 antibody and the drug monomethyl auristatin E (MMAE). It is an anti-neoplastic agent used in the treatment of Hodgkin lymphoma and systemic anaplastic large cell lymphoma and was approved in 2011. In January 2012, the drug label was revised to include a boxed warning of progressive multifocal leukoencephalopathy and death following JC virus infection. The structure of the cAC10-vcMMAE system used to produce brentuximab vedotin is illustrated in figure 3.
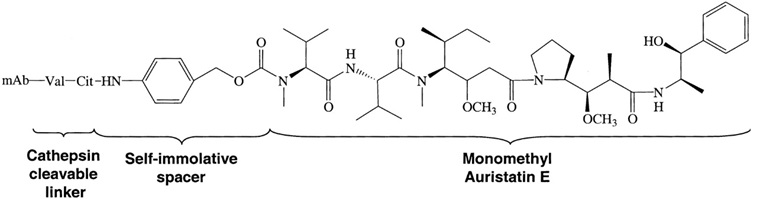
Figure 3: Structure of the cAC10-vcMMAE system used to produce brentuximab vedotin. Francisco et al. in 2003 report how this ADC was prepared. A controlled partial reduction of internal cAC10 disulfides with DTT, followed by addition of the maleimide-vc-linker-MMAE was used. The stable thioether-linked ADCs were formed by the addition of free sulfhydryl groups on the mAbs to the maleimides present on the drugs. cAC10-vcMMAE and the cIgG-vcMMAE used in the report contained approximately 8 drugs/mAb.
The development of ADCs in the past decade has evolved from murine antibodies to conjugate standard chemotherapeutics drugs to fully human antibodies conjugated to highly potent cytotoxic drugs. Critical factors required for the successful development of ADCs, as scientists have learned over the past decade, include target antigen selection and the selection of the best antibody, the correct linker, and payload. Typically, ADCs are complex biomolecules composed of an antibody or antibody fragment linked to a biologically active cytotoxic or anti-cancerous payload using stable chemical linkers with labile bonds. Other terms used for this molecule are “bioconjugate” and “immune-conjugates.” Combining the unique targeting abilities of antibodies with cancer-killing drugs to create unique ADCs is the potential these drugs can have to treat cancer that is hard to treat with standard classical chemotherapies.
One area of research essential for developing and designing ADCs is conjugation chemistry. One recent improvement is the implementation of site-specific conjugation methods. Here, the conjugation only occurs at engineered cysteine residues or unnatural amino acids. The result is a homogeneous ADC production and improved ADC pharmacokinetic (PK) properties. The following figure illustrates critical factors that influence the performance of ADC therapeutics.
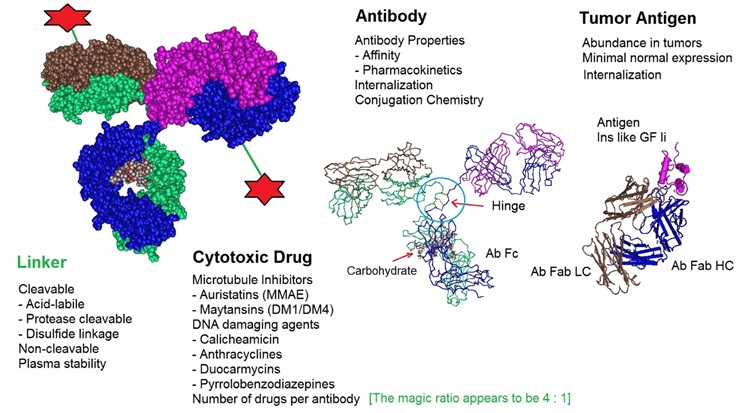
Figure 4: Critical factors that influence Antibody Drug Conjugate or ADC therapeutics. ADCs are designed and produced by conjugating a cytotoxic drug to a monoclonal antibody via a selected linker. All components of an ADC affect the performance of the molecule. The optimization of all molecular parts is essential for the development of successful ADCs.
Reference
Argos P. An investigation of oligopeptides linking domains in protein tertiary structures and possible candidates for general gene fusion. J Mol Biol. 1990;211:943–958.
Chen X, Bai Y, Zaro J, Shen WC. Design of an in vivo cleavable disulfide linker in recombinant fusion proteins. Biotechniques. 2010;49:513–518.
Chen X, Zaro JL, Shen WC. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev. 2013 Oct;65(10):1357-69. [PMC]
Franco Dosio, Paola Brusa and Luigi Cattel; Immunotoxins and Anticancer Drug Conjugate Assemblies: The Role of the Linkage between Components. Toxins 2011, 3, 848-883. [PMC]
Ducry L, Stump B.; Antibody-drug conjugates: linking cytotoxic payloads to monoclonal antibodies. Bioconjug Chem. 2010 Jan;21(1):5-13.
Feng Tiana, Yingchun Lua, Anthony Manibusana, Aaron Sellersa, Hon Trana, Ying Suna, Trung Phuonga, Richard Barnetta, Brad Hehlia, Frank Songa, Michael J. DeGuzmanb, Semsi Ensarib, Jason K. Pinkstaffc, Lorraine M. Sullivanc, Sandra L. Birocc, Ho Chod, Peter G. Schultze, John DiJoseph, Maureen Dougher, Dangshe Ma, Russell Dushing, Mauricio Lealh, Lioudmila Tchistiakovai, Eric Feyfanti, Hans-Peter Gerber, and Puja Sapra; A general approach to site-specific antibody drug conjugates. (2014) PNAS 111, 5, 1766-1771.
Francisco, J,A, Charles G. Cerveny, Damon L. Meyer, Bruce J. Mixan, Kerry Klussman, Dana F. Chace, Starr X. Rejniak, Kristine A. Gordon, Ron DeBlanc, Brian E. Toki, Che-Leung Law, Svetlana O. Doronina, Clay B. Siegall, Peter D. Senter, and Alan F. Wahl; cAC10-vcMMAE, an anti-CD30–monomethyl auristatin E conjugate with potent and selective antitumor activity. August 15, 2003; Blood: 102 (4).
George R, & Heringa J. An analysis of protein domain linkers: their classification and role in protein folding. Protein Eng. 2002;15:871–879.
SEAN L KITSON, DEREK J QUINN, THOMAS S MOODY, DAVID SPEED, WILLIAM WATTERS, DAVID ROZZELL; Antibody-Drug Conjugates (ADCs) – Biotherapeutic bullets. Chemistry Today - vol. 31(4) July/August 2013.
Leung MR, van Bezouwen LS, Schopfer LM, Sussman JL, Silman I, Lockridge O, Zeev-Ben-Mordehai T. Cryo-EM structure of the native butyrylcholinesterase tetramer reveals a dimer of dimers stabilized by a superhelical assembly. Proc Natl Acad Sci U S A. 2018 Dec 26;115(52):13270-13275. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6310839/
Lewis Phillips GD, Li G, Dugger DL, Crocker LM, Parsons KL, Mai E, Blättler WA, Lambert JM, Chari RV, Lutz RJ, Wong WL, Jacobson FS, Koeppen H, Schwall RH, Kenkare-Mitra SR, Spencer SD, Sliwkowski MX; Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res. 2008 Nov 15;68(22):9280-90.
McCombs JR, Owen SC. Antibody drug conjugates: design and selection of linker, payload and conjugation chemistry. AAPS J. 2015 Mar;17(2):339-51. Epub 2015 Jan 22. PMID: 25604608; PMCID: PMC4365093. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4365093/
Mottram JC, North MJ, Barry JD, Coombs GH. A cysteine proteinase cDNA from Trypanosoma brucei predicts an enzyme with an unusual C-terminal extension. FEBS Lett. 1989 Dec 4;258(2):211-5. doi: 10.1016/0014-5793(89)81655-2. PMID: 2599086. A cysteine proteinase cDNA from Trypanosoma brucei predicts an enzyme with an unusual C‐terminal extension (wiley.com)
Nilvebrant J, Åstrand M, Georgieva-Kotseva M, Björnmalm M, Löfblom J, Hober, S.; (2014) Engineering of Bispecific Affinity Proteins with High Affinity for ERBB2 and Adaptable Binding to Albumin. PLoS ONE 9(8): e103094. doi:10.1371/journal.pone.0103094.
Panowksi S, Bhakta S, Raab H, Polakis P, Junutula JR.; Site-specific antibody drug conjugates for cancer therapy. MAbs. 2014 Jan-Feb;6(1):34-45. doi: 10.4161/mabs.27022.
Sassoon I, Blanc V.; Antibody-drug conjugate (ADC) clinical pipeline: a review. Methods Mol Biol. 2013;1045:1-27. doi: 10.1007/978-1-62703-541-5_1.
Turner S, Russell G, Williamson M, Guest J. Restructuring an interdomain linker in the dihydrolipoamide acetyltransferase component of the pyruvate dehydrogenase complex of Escherichia coli. Protein Eng. 1993;6:101–108.
Whitley MJ, Arjunan P, Nemeria NS, Korotchkina LG, Park YH, Patel MS, Jordan F, Furey W. Pyruvate dehydrogenase complex deficiency is linked to regulatory loop disorder in the αV138M variant of human pyruvate dehydrogenase. J Biol Chem. 2018 Aug 24;293(34):13204-13213. doi: 10.1074/jbc.RA118.003996. Epub 2018 Jul 3. PMID: 29970614; PMCID: PMC6109939. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6109939/
Zhao HL, Xue C, Du JL, Ren M, Xia S, Liu ZM. Balancing the pharmacokinetics and pharmacodynamics of interferon-alpha2b and human serum albumin fusion protein by proteolytic or reductive cleavage increases its in vivo therapeutic efficacy. Mol Pharm. 2012;9:664–670.
---...---
Bio-Synthesis provides a full spectrum of bio-conjugation services including high quality custom oligonucleotide modification services, back-bone modifications, conjugation to fatty acids and lipids, cholesterol, tocopherol, peptides as well as biotinylation by direct solid-phase chemical synthesis or enzyme-assisted approaches to obtain artificially modified oligonucleotides, such as BNA antisense oligonucleotides, mRNAs or siRNAs, containing a natural or modified backbone, as well as base, sugar and internucleotide linkages.
Bio-Synthesis also provides biotinylated mRNA and long circular oligonucleotides.
---...---