
Bio-Synthesis Inc. is pleased to offer a large variety of RNA molecules (both synthetic and enzymatically derived) for a number of research applications, including COVID 19 testing and analysis !
We also have COVID19 RNA controls such as those currently being used by FDA approved laboratories for COVID molecular testing !
The coronavirus SARS-CoV in 2002 to 2003 caused an outbreak of severe acute respiratory syndrom (SARS). Similar to SARS-CoV, the new SARS-CoV-2 (COVID-19) virus causes a zoonotic infection of the respiratory system in humans. This new virus is related to the SARS-associated coronavirus (SARS-CoV), however it is not the same virus.
According to the CDC, SARS was first discovered in Asia in February 2003. The outbreak lasted approximately six months. SARS-CoV 2003 spread to more than two dozen countries in North America, South America, Europe, and Asia before it stopped in July 2003.
A characteristic feature of coronavirus genomes is that approximately two-thirds of their (+)-sense RNA genome code for overlapping replicase genes ORF 1a and ORF 1 ab. The other one-third encodes a set of subgenomic mRNAs required for accessory proteins and structural proteins.
According to Sawicki et al. (2007), the virus genome encodes structural proteins, as well as nonstructural proteins. Structural proteins are critical in viral RNA synthesis and referred to as replicase-transcriptase proteins. Non-structural proteins are thought as nonessential for virus replication in cell culture. These nonessential proteins appear to give the virus a selective advantage in vivo and are considered as niche-specific proteins. The niche-specific protein, nonstructural protein 2 (nsp2) together with the structural protein N, the nucleocapsid protein, take part in viral RNA synthesis.
Krichel et al. in 2020 studied the processing of the SARS-CoV pp1a/ab nsp7-10 region and reported that initially, ORF 1a and ORF 1ab are directly translated into either replicase poly protein pp1a (nsp1 to 11) or pp1ab (nsp1to 16), respectively. A ribosomal (-1)-frameshift is responsible for differential translation of ORF 1a and ORF 1ab. Figure 1 illustrates the organization of the SARS-CoV-2 genome as well as the structural relationships of the genome- and subgenome-length mRNAs.
.jpg)
Figure 1: Organization of the SARS-CoV-2 genome. The structural relationship of the genome- and subgenome-length mRNAs is illustrated. The proteolytic processing of the replicase polyprotein 1ab is shown in the left part of the figure. [Genome]
Replicase Polyprotein 1ab
.jpg)
Figure 2: Features of the replicase polyprotein 1ab [Source: P0C6X7.1].
The translated polyproteins undergo proteolytic processing into 11 or 16 individual nsp’s. The nsp’s are part of the replication/transcription complex (RTC). The RTC is a membrane-anchored, highly dynamic protein-RNA complex needed for the replication processes. Biochemical studies that included subcellular fractionation, in-situ hybridization observed with electron microscopy combined immunofluorescence suggest that most, if not all, coronavirus nsp proteins are part of the RTC. The RTC synthesizes both genome- and subgenome-length RNA.
Two coronavirus proteases are known to facilitate processing of the polyprotein. A papain-like protease (PLpro; nsp3) encoded between nsp 1 to 4, and the main chymotrypsin-like protease (Mpro; 3CLpro, nsp5) encoded between nsp 4 to 11/16. These two proteases are essential. Therefore they have already been heavily investigated. Scientist studied their structure and function for suitability as drug targets.
Maturation of the polyprotein via auto-processing of nsp 4-5 and nsp 5-6 regulates the activity of MPro protease, as well as concentration and substrate-induced self-assembly into an active dimeric unit. The freed MPro protease liberates nsp’s 6 to 16 from the polyprotein by targeting the nsp inter-domain junctions. These junctions contain primarily -LQꜜS- or -LQꜜA- amino acid residues at positions P2, P1 and P1’. However, it appears that only Q at P1 is needed.
Krichel et al. studied polyprotein maturation of SARS-CoV using cloning, expression, and purification of the His-tagged CoV ORF1ab nsp 7-10 region. For protein production, the researchers use an auto-cleavable GST-tag in combination with a His-tag. To monitor protein processing, a FRET peptide assay, SDS-PAGE, and native mass spectrometry were employed to elucidate specific substrate efficiencies as well as protein processing pathways.
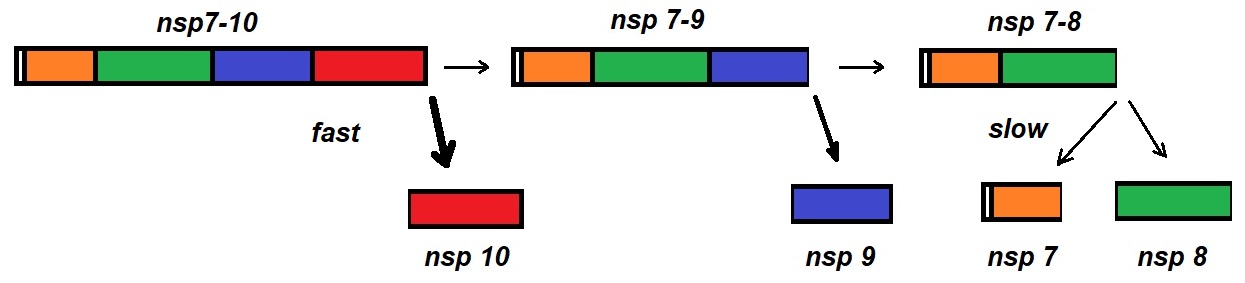
Figure 3: SARS-CoV nsp 7-10 processing. Krichel et al. utilized native mass spectrometry for monitoring the processing of the polyprotein region. This figure illustrates the observed cleavage order and efficiency.
Reference
Coronavirus 2019
SARS History
Krichel B, Falke S, Hilgenfeld R, Redecke L, Uetrecht C.; Processing of the SARS-CoV pp1a/ab nsp7-10 region. Biochem J. 2020 Mar 13;477(5):1009-1019. [PMC]
Replicase polyprotein 1ab
Sawicki SG, Sawicki DL, Siddell SG.; A contemporary view of coronavirus transcription. J Virol. 2007 Jan;81(1):20-9. doi: 10.1128/JVI.01358-06. Epub 2006 Aug 23. PMID: 16928755; PMCID: PMC1797243. [PMC]
---...---