Coronaviruses (CoVs) are enveloped positive-sense RNA viruses. The club-like spikes projecting out from their surface gave them the name. Coronaviruses possess an unusual large RNA genome as well as a unique replication strategy. Coronaviruses cause a variety of diseases in animals ranging from cows, pigs to chicken, and other birds. In humans, coronaviruses can cause potentially lethal respiratory infections.
Coronaviruses belong to the largest group of viruses called the Nidovirales order. Members of this order include the Coronaviridae, Arteriviridae, and Roniviridae families. The Coronvirinae are one of two subfamilies in the Coronaviridae family. Coronavirinae are further subdivided into for groups, the alpha, beta, gamma, and delta coronaviruses. Nowadays, these viruses are divided using phylogenetic clustering. These virus families have animal and human hosts. The Middle Eastern Respiratory Syndrome Coronavirus (MERS-CoV) and Severe Acute Respiratory Coronavirus (SARS-CoV) are examples.
Nidoviruses contain an infectious, linear, positive-sense RNA genome that is capped and polyadenylated. Based on their genome size, nidoviruses are divided into two groups large and small nidoviruses.
All Nidovirales viruses are enveloped, non-segmented positive-sense RNA viruses containing very huge genomes.
Common features of coronaviruses include
(i) a highly conserved genomic organization with a large replicase gene preceding structural and accessory genes,
(ii) expression of many non-structural genes by ribosomal frameshifting,
(iii) several unique of unusual enzymatic activities encoded within the large replicase-transcriptase polyprotein, and
(iv) expression of downstream genes by synthesis of 3’-nested sub-genomic mRNAs.
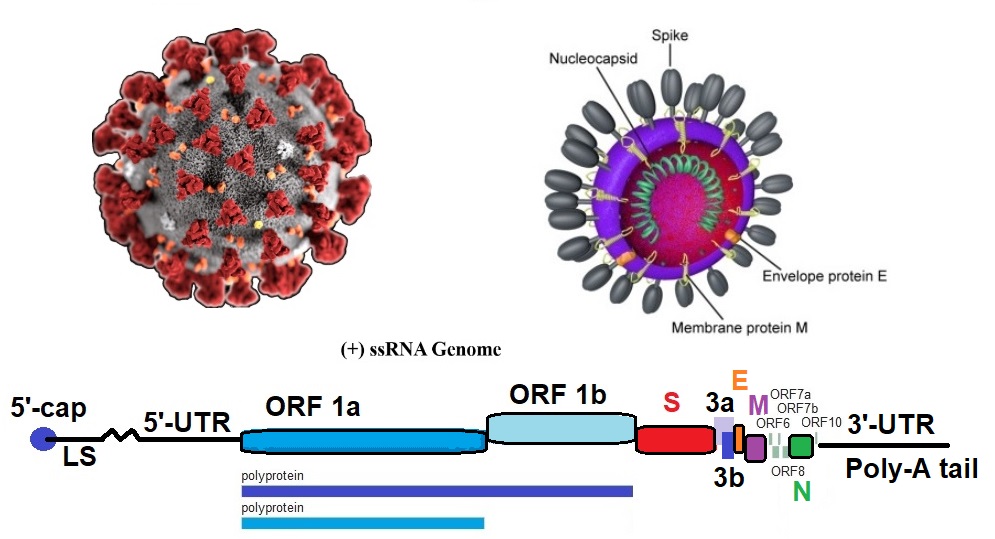
The typical organization of the genome is
5’-leader-UTR-replicase-S(Spike)-E(Envelope)-M(Membrane)-N(Nucleocapsid)-3’-UTR-poly(A) tail.
Accessory genes are interspersed within the structural genes at the 3’-end of the genome.
Accessory proteins are not needed for replication in tissue culture but appear to be important in viral pathogenesis. The synthesis of polypeptide 1ab (pp1ab) involves programmed ribosomal frame shifting during translation of open-reading frame 1a (orf1a). Frame shifting results in a new reading frame that produces a trans-frame protein product. In coronaviruses, a fixed portion of the ribosomes translating orf1a change reading frame at a specific location now decoding information contained in orf1b.
U_UUA_AAC is a universal frame-shifting site
Coronaviruses contain a frameshifting stimulation element as a conserved RNA sequence forming a stem-loop that promotes ribosomal frameshifting. Ribosomal frameshifting is a mechanism in which open-reading frame 1b (orf1b) is expressed. Replicase-transcriptase proteins are encoded in open-reading frame 1a and 1b (orf1a and orf1b) and are synthesized initially as two large polyproteins termed pp1a and pp1b. A comparative analysis performed by Baranov et al. in 2004 revealed the sequence U_UUA_AAC as a universal shift site. Frameshifting was characterized in SARS-CoV cultured in mammalian cells using a dual luciferse reporter system and mass spectrometry. Tandem tRNA slippage on the sequence U_UUA_AAC was confirmed by mutagenic analysis of the shift site. Mass spectrometry was used for the analysis of affinity tagged frameshift products. Further analysis of the frameshifting site showed that a proposed RNA secondary structure in loop II and two unpaired nucleotides at the stem I-stem II junction in SARS-CoV are important for frameshift stimulation.
SARS-CoV-2 (COVID-19)
Taxonomy: Group IV ((+)single-stranded RNA, ssRNA); Coronviridae; Coronavirinae; Betacoronavirus;
Sarbecovirus; Severe Acut Respiratory Cornavirus 2 (SARS-CoV-2)
Virion: Enveloped, sperical, 60 to 140 nm in diameter with 9 to 12 nm spikes
Genome: ~ 30 kb positive-sense, ssRNA
RNA Transcript: 5'-cap, 3'-poly-A tail
Proteome: 10 proteins
Transmission: Links to seafood and animal market cases suggest animal-to-human transmission.
Sustained human-to-human transmission observed in later cases.
Phylogeny: Closely related to bat-SL-CoVZC45 and bat-SL-CoVZX21.
Reference Genome: GenBank: MN908947; PMID: 31978945; CDC-2019-nCoV
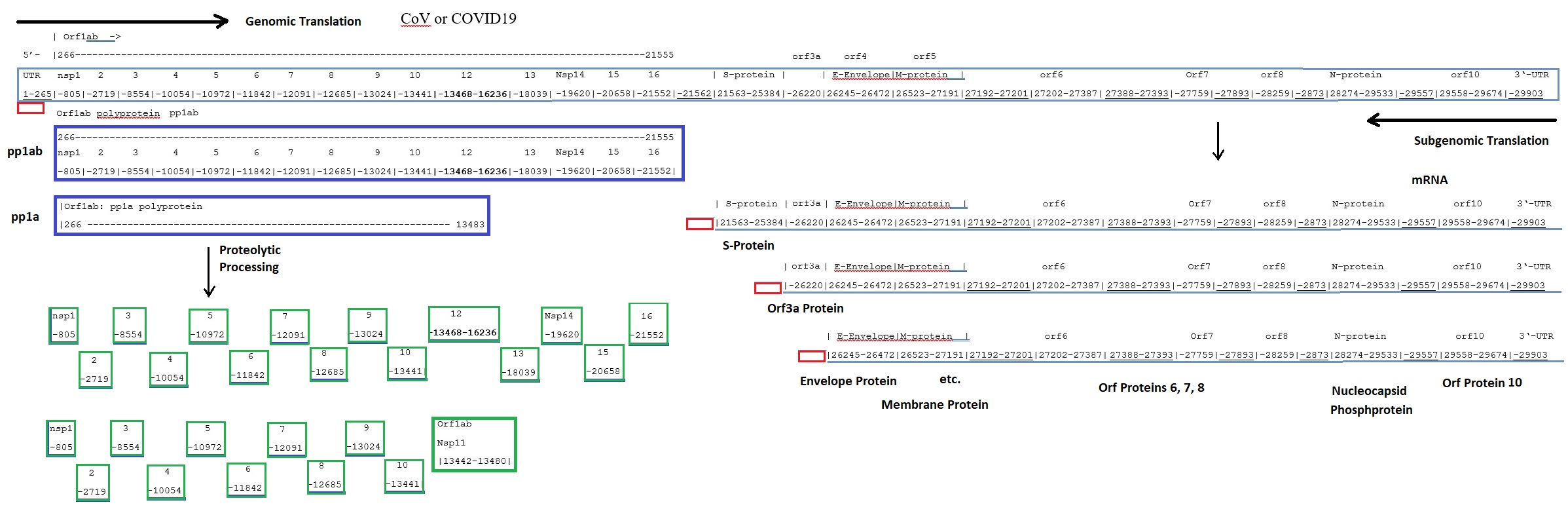
Model of Coronavirus COVID19 Transcription. A possible model of the coronavirus COVID19 transcription mechanism is shown here. The model is based on the genomic sequence and the model for the transcription of coronaviruses as proposed by Sawicki et al. in 2007. The organization and the expression of the Wuhan seafood market pneumonia virus isolate [reference genome] Wuhan-Hu-1 genome is depicted here. Structural relationships of the genome and subgenome mRNAs are shown. Orfs are defined by the published genome sequence. Possible autoproteolytic processing of orfs1a and orf1ab polypeptides into protein nsp1 to 16 are shown as well.
Reference
Baranov PV, Henderson CM, Anderson CB, Gesteland RF, Atkins JF, Howard MT (February 2005). "Programmed ribosomal frameshifting in decoding the SARS-CoV genome". Virology. 332 (2): 498-510. [Pubmed]
Buchan, J.R.; Stansfield, I. (2007). "Halting a cellular production line: responses to ribosomal pausing during translation". Biol Cell. 99 (9): 475–487. [Source]
Fehr & Perlman; Coronaviruses: An overview of their replication and pathogenesis. Method Mol Biol. 2015; 1282:1-23. [PMC]
Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. J Virol. 2007 Jan;81(1):20-9. doi: 10.1128/JVI.01358-06. Epub 2006 Aug 23. PMID: 16928755; PMCID: PMC1797243. [PMC]
Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z; The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U.S.A. (2003) 100 p.13190-5. [Pubmed]
Genomic structure of Wuhan seafood market pneumonia virus [Now COVID-19]
Isolate 2019-nCoV/USA-AZ1/2020 - 2019 Outbreak Info
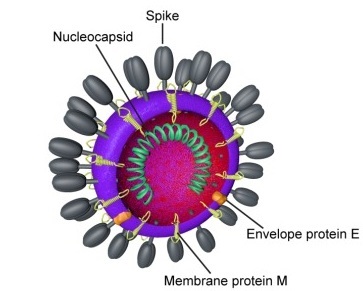
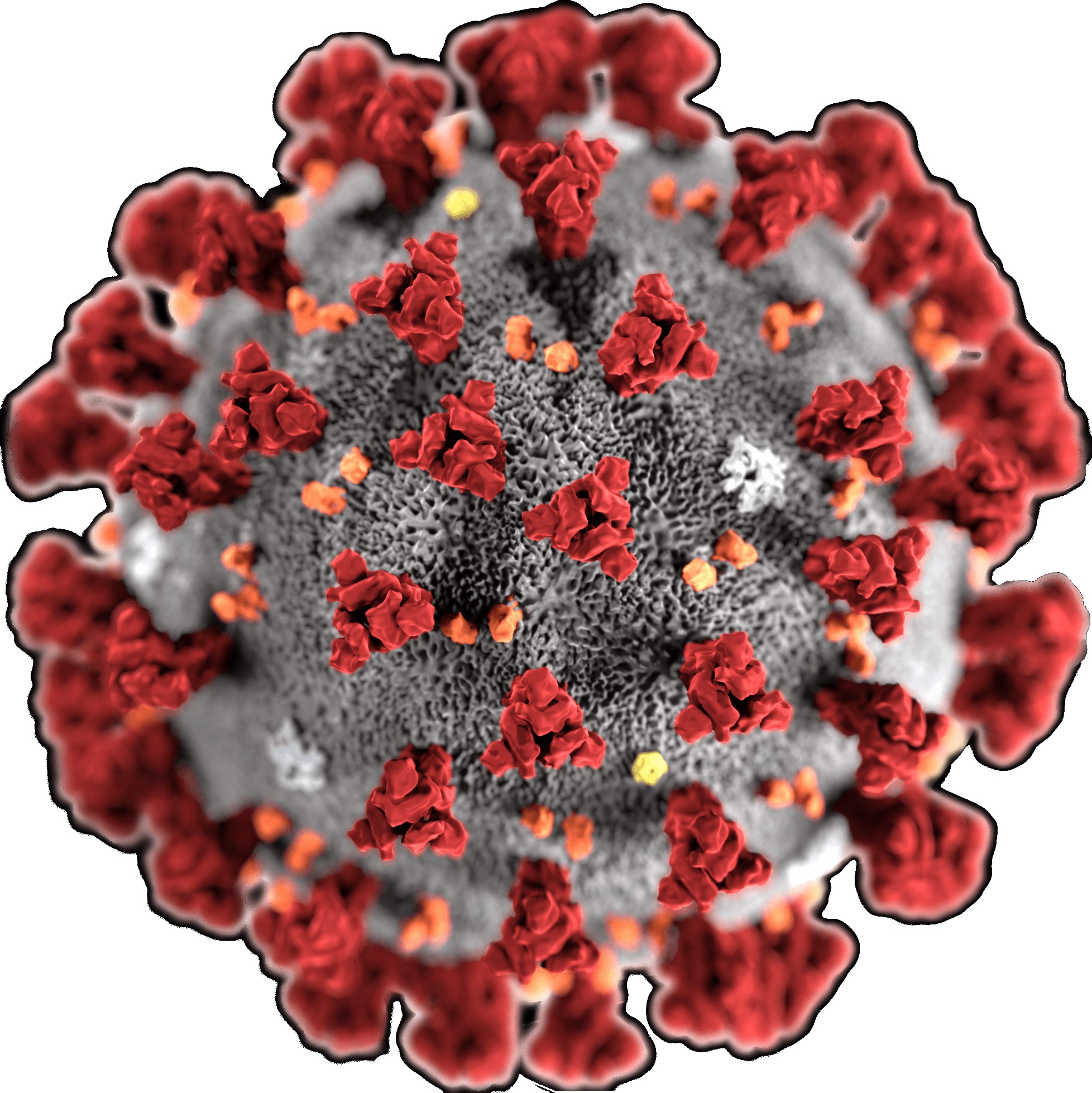
Source: Wiki Commonns; CDC Commons
|
#
|
Position
|
1..29903 WSMPV Wuhan seafood market pneumonia virus
|
|
1
|
1-265
|
5’-UTR
|
|
2
|
266-21555
|
Orf1ab: Polyprotein. Ribosomal slippage, id "QHQ82463.1"
|
|
3
|
266-805
|
Orf1ab; nsp1: Leader protein produced by both pp1a and pp1ab.
Protein id "YP_009725297.1" .
Promotes cellular mRNA degradation and blocks host cell translation.
The result is blocking of the innate immune response.
|
|
4
|
806-2719
|
Orf1ab: nsp2, produced by both pp1a and pp1ab. protein_id="YP_009725298.1".
Binds to prohibitin proteins. No known function as of 2020 (?).
|
|
5
|
2720-8554
|
Orf1ab: nsp3: Contains conserved domains: N-terminal acidic (Ac), predicted phosphoesterase, papain-like proteinase, Y-domain, transmembrane domain 1 (TM1), adenosine diphosphate-ribose 1''-phosphatase (ADRP); produced by both pp1a and pp1ab.
Protein id "YP_009725299.1".
Large, multidomain transmembrane protein. Activities include:
a) ubiquitin-like 1 (Ubl1) and Ac domains interacting with N protein;
b) ADRP activity promoting cytokine expression;
c) Papain-like protease (PLPro)/Deubiquitinase domain.
Cleaves viral polyprotein and blocks host immune response.
Ubiquitin-like 2 (UBl2), nucleic acid binding (NAB), G2M,
SARS-unique domain (SUD), Y domains of unknown function.
Known structures:
https://www.ncbi.nlm.nih.gov/structure/?term=SARS-CoV+PLpro
|
|
6
|
8555-10054
|
Orf1ab: nsp4, contains transmembrane domain 2 (TM2);
produced by both pp1a and pp1ab.
Protein id "YP_009725300.1".
Potential transmembrane scaffold protein, important for proper structure of double-membrane vesicles (DMVs).
|
|
7
|
10055-10972
|
Orf1ab: 3C-like proteinase; nsp5: Main proteinase (Mpro). Mediates cleavages downstream of nsp4.
The 3D structure [1UK3] for the severe acute respiratory syndrome (SARS) virus main protease has been determined (Yang et al., 2003); produced by both pp1a and pp1ab.
.jpg)
Protein_id "YP_009725301.1". Cleaves viral polyprotein.
>pdb|1UK3|A Chain A, Crystal Structure Of Sars Coronavirus Main Proteinase (3clpro) At Ph7.6
1--------10--------20--------30--------40--------50
SGFRKMAFPSGKVEGCMVQVTCGTTTLNGLWLDDTVYCPRHVICTAEDML
NPNYEDLLIRKSNHSFLVQAGNVQLRVIGHSMQNCLLRLKVDTSNPKTPK
YKFVRIQPGQTFSVLACYNGSPSGVYQCAMRPNHTIKGSFLNGSCGSVGF
NIDYDCVSFCYMHHMELPTGVHAGTDLEGKFYGPFVDRQTAQAAGTDTTI
TLNVLAWLYAAVINGDRWFLNRFTTTLNDFNLVAMKYNYEPLTQDHVDIL
GPLSAQTGIAVLDMCAALKELLQNGMNGRTILGSTILEDEFTPFDVVRQC
SGVTFQ
|
|
8
|
10973-11842
|
Orf1ab: nsp6; putative transmembrane domain; produced by both pp1a and pp1ab. Protein id "YP_009725302.1".
|
|
9
|
11843-12091
|
Orf1ab: nsp7; produced by both pp1a and pp1ab. Protein id "YP_009725303.1".
Forms a hexadecameric complex with nsp8 and may act as a processivity clamp for RNA polymerase.
Structures for SARS-CoV nsp12-nsp7-nsp8 cofactors are known.
|
|
10
|
12092-12685
|
Orf1ab: nsp8; produced by both pp1a and pp1ab. Protein id "YP_009725304.1".
Forms a hexadecameric complex with nsp7 and may act as processivity clamp for RNA polymerase and/or primase.
|
|
11
|
12686-13024
|
Orf1ab: nsp9; ssRNA-binding protein; produced by both pp1a and pp1ab.
Protein id "YP_009725305.1".
|
|
12
|
13025-13441
|
Orf1ab: nsp10; formerly known as growth-factor-like protein (GFL). Produced by both pp1a and pp1ab.
Protein id "YP_009725306.1". Cofactor for nsp16 and nsp14.
Forms a heterodimer with both and stimulates viral exoribonuclease (ExoN) and 2-O-methyltransferase
(2-O-MT) activity.
|
|
13
|
13442-13468,
13468-16236
|
Orf1ab: RNA-dependent RNA polymerase; nsp12;
RNA-dependent RNA-polymerase (RdRp).
Produced by pp1ab only. Protein id "YP_009725307.1".
|
|
14
|
16237-18039
|
Orf1ab: helicase; nsp13; zinc-binding domain (ZD), NTPase/helicase domain (HEL), RNA 5'-triphosphatase; produced by pp1ab only.
Protein id "YP_009725308.1".
|
|
15
|
18040-19620
|
Orf1ab: 3'-to-5' exonuclease; nsp14; produced by pp1ab only.
Protein id "YP_009725309.1.
N7 methyl-transferase (MTase) and 3‘-5‘-exoribonuclease (ExoN).
ExoN activity is important for proofreading of viral genome.
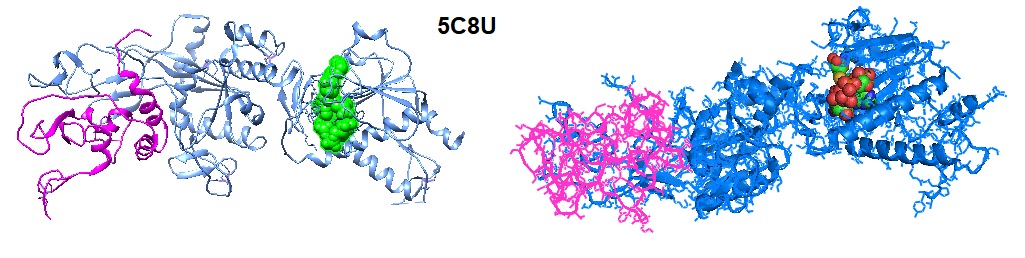
Cartoon representation of the structure of the nsp14–nsp10 heterodimer.
|
|
16
|
19621-20658
|
Orf1ab: endoRNAse; nsp15; produced by pp1ab only.
Protein id "YP_009725310.1".
Viral endoribonuclease (NendoU). A structure for the nsp15 (F307L) protein from the MHV coronavirus
was solved in 2006.
|
|
17
|
20659-21552
|
Orf1ab: 2'-O-ribose methyltransferase; nsp16; 2'-O-MT; produced by pp1ab only.
Protein id "YP_009725311.1".
|
|
18
|
266-13483
|
Orf1ab: pp1a; orf1a polyprotein.
Protein id "YP_009725295.1". GeneID:"43740578".
2’-O-MT shielding viral RNA from Melanoma differentiation associated protein 5 (mMDA5) recognition.
|
|
19
|
13442-13480
|
Orf1ab: nsp11; produced by pp1a only". Protein_id="YP_009725312.1".
|
|
20
|
21563-25384
|
S gene = Surface glycoprotein. "QHQ82464.1"
S: Structural protein; spike protein.
Protein id "YP_009724390.1"; GeneID: "43740568"
|
|
21
|
25393-26220
|
orf3a, “orf3a protein",”QHQ82465.1".
1--------10--------20--------30--------40--------50
MDLFMRIFTIGTVTLKQGEIKDATPSDFVRATATIPIQASLPFGWLIVGV
ALLAVFQSASKIITLKKRWQLALSKGVHFVCNLLLLFVTVYSHLLLVAAG
LEPFLYLYALVYFLQSINFVRIIMRLWLCWKCRSKNPLLYDANYFLCWHT
NCYDYCIPYNSVTSSIVITSGDGTTSPISEHDYQIGGYTEKWESGVKDCV
VLHSYFTSDYYQLYSTQLSTDTGVEHVTFFIYNKIVDEPEEHVQIHTIDG
SSGVVNPVMEPIYDEPTTTTSVPL
|
|
22
|
26245-26472
|
E gene = Envelope Protein "QHQ82466.1"
1--------10--------20--------30--------40--------50
MYSFVSEETGTLIVNSVLLFLAFVVFLLVTLAILTALRLCAYCCNIVNVSL
VKPSFYVYSRVKNLNSSRVPDLLV
|
|
23
|
26523-27191
|
M gene: ORF5; structural protein. start=1.
Membrane glycoprotein.
Protein_id “YP_009724393.1”. GeneID “43740571"
1--------10--------20--------30--------40--------50
MADSNGTITVEELKKLLEQWNLVIGFLFLTWICLLQFAYANRNRFLYIIKL
IFLWLLWPVTLACFVLAAVYRINWITGGIAIAMACLVGLMWLSYFIASFRL
FARTRSMWSFNPETNILLNVPLHGTILTRPLLESELVIGAVILRGHLRIAG
HHLGRCDIKDLPKEITVATSRTLSYYKLGASQRVAGDSGFAAYSRYRIGNY
KLNTDHSSSSDNIALLVQ
|
|
24
|
27192-27201
|
?
|
|
25
|
27202-27387
|
Orf6; Protein id "QHQ82468.1.“.
1--------10--------20--------30--------40--------50
MFHLVDFQVTIAEILLIIMRTFKVSIWNLDYIINLIIKNLSKSLTENKYSQ
LDEEQPMEID
ORF6; protein id "YP_009724394.1“. GeneID “43740572"
1--------10--------20--------30--------40--------50
MFHLVDFQVTIAEILLIIMRTFKVSIWNLDYIINLIIKNLSKSLTENKYSQ
LDEEQPMEID
|
|
26
|
27387-27393
|
?
|
|
27
|
27394-27759
|
ORF7a: GeneID “43740573”. ORF7a protein. Protein_id id "YP_009724395.1”.
1--------10--------20--------30--------40--------50
MKIILFLALITLATCELYHYQECVRGTTVLLKEPCSSGTYEGNSPFHPLA
DNKFALTCFSTQFAFACPDGVKHVYQLRARSVSPKLFIRQEEVQELYSPI
FLIVAAIVFITLCFTLKRKTE
Protein id="YP_009724395.1. GeneID:43740573
MKIILFLALITLATCELYHYQECVRGTTVLLKEPCSSGTYEGNSPFHPLA
DNKFALTCFSTQFAFACPDGVKHVYQLRARSVSPKLFIRQEEVQELYSPI
FLIVAAIVFITLCFTLKRKTE
|
|
28
|
27894-28259
|
ORF8: ORF8 protein. Protein id "YP_009724396.1. GeneID:”43740577".
MKFLVFLGIITTVAAFHQECSLQSCTQHQPYVVDDPCPIHFYSKWYIRVG
ARKSAPLIELCVDEAGSKSPIQYIDIGNYTVSCLPFTINCQEPKLGSLVV
RCSFYEDFLEYHDVRVVLDFI
|
|
29
|
28274-29533
|
N Protein: ORF9; structural protein. Nucleocapsid phosphoprotein.
Protein id "YP_009724397.2”. GeneID: ”43740575”.
1--------10--------20--------30--------40--------50
MSDNGPQNQRNAPRITFGGPSDSTGSNQNGERSGARSKQRRPQGLPNNTA
SWFTALTQHGKEDLKFPRGQGVPINTNSSPDDQIGYYRRATRRIRGGDGK
MKDLSPRWYFYYLGTGPEAGLPYGANKDGIIWVATEGALNTPKDHIGTRN
PANNAAIVLQLPQGTTLPKGFYAEGSRGGSQASSRSSSRSRNSSRNSTPG
SSRGTSPARMAGNGGDAALALLLLDRLNQLESKMSGKGQQQQGQTVTKKS
AAEASKKPRQKRTATKAYNVTQAFGRRGPEQTQGNFGDQELIRQGTDYKH
WPQIAQFAPSASAFFGMSRIGMEVTPSGTWLTYTGAIKLDDKDPNFKDQV
ILLNKHIDAYKTFPPTEPKKDKKKKADETQALPQRQKKQQTVTLLPAADL
DDFSKQLQQSMSSADSTQA
|
|
30
|
29558-29674
|
ORF10: ORF10 protein. Protein_id "YP_009725255.1"
GeneID "43740576"
MGYINVFAFPFTIYSLLLCRMNSRNYIAQVDVVNFNLT
|
|
31
|
29675-29903
|
3‘-UTR
|
---...---
" Bio-Synthesis provides a full spectrum of high quality custom oligonucleotide modification services including back-bone modifications, conjugation to fatty acids, biotinylation by direct solid-phase chemical synthesis or enzyme-assisted approaches to obtain artificially modified oligonucleotides, such as BNA antisense oligonucleotides, mRNAs or siRNAs, containing a natural or modified backbone, as well as base, sugar and internucleotide linkages.
Bio-Synthesis also provides biotinylated mRNA and long circular oligonucleotides".
---...---