The new coronavirus (2019-nCoV) first detected in Wuhan City, Hubei Province, China, has infected tens of thousands of people in China. The virus is spreading from person-to-person in almost all parts of the world. The alarming fast transmission of this virus resulted in the World Health Organization (WHO) declaring a global health emergency on 30 January 2020. As of April 29, 2002, a total of 981,246 infections and 55,258 deaths have been reported in the US alone.
Coronaviruses are a large family of viruses that are common in many animals, including camels, cattle, cats, and bats. However, some animal coronaviruses can infect people and spread between people, for example, MERS, SARS, and now 2019-nCoV (or SARS-CoV-2 or COV19) {CDC}.
Xu et al. recently (in February 2020) compared the sequences of animal-to-human transmitted Human Coronaviruses: SARS-CoV-2, SARS-CoV and MERS-CoV.
SARS-CoV-2 was isolated from the respiratory epithelium of a patient with unexplained pneumonia symptoms in the Wuhan seafood market where the coronavirus outbreak occurred. The genomic comparison revealed that the genomes of SRA-Cov-2 and SARS-CoV are extremely homologous at the nucleotide level. Xu et al. identified six regions of difference (RD) in the genome sequence between SARS-CoV and SRSR-CoV-2. RD1, 448 nucleotides in size, RD2, 55 nucleotides in size, and RD3, 278 nucleotides in size, are partial coding sequences of the orf 1ab gene. RD4, 315 nucleotides long, and RD5, 80 nucleotides long, are partial coding sequences of the S gene. RD6, 214 nucleotides in size, is part of the coding sequence of the orf7b and orf8 genes.
Megha Satyanarayana reported in CEN March 2/9 2020 that the CDC’s coronavirus test kit used for the diagnosis of the SARS-CoV-2 was plagued with problems. The accuracy of the testing kit was cast into doubt since it gave inconclusive readouts. According to CDC officials, positive SARS-CoV-2 tests at the state level must be confirmed by the CDC. Therefore, the careful design and validation of all probes used for the detection of RNA sequences of all selected coronaviruses is a necessity. Positive and negative controls will also need to work accurately. Hence, there appears to be a need for primer and probe sets that accurately detect the newly emerged coronavirus SARS-CoV-2 (COV19).
Update: CDC COVID-19: SARS-CoV-2 Variant Classifications and Definitions
Reference
Xu J, Zhao S, Teng T, Abdalla AE, Zhu W, Xie L, Wang Y, Guo X. Systematic Comparison of Two Animal-to-Human Transmitted Human Coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020 Feb 22;12(2):244. [PMC]
Corona virus outbreaks in January 2020.
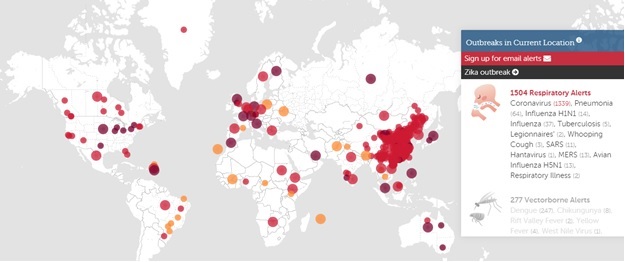
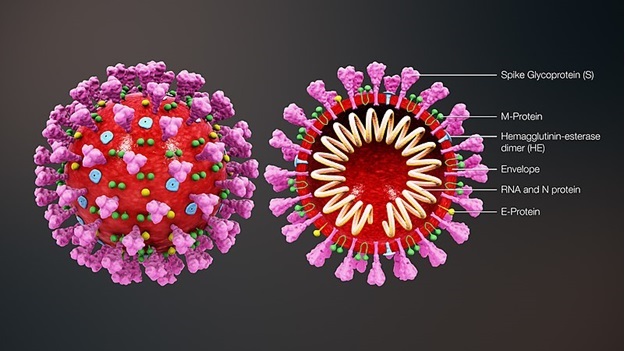
Source: Wikimedia Commons
Probes are labeled with a reporter fluorophore such as 6-carboxyfluorescein (FAM), usually on the 5'-end, and a quencher molecule, such as carboxytetramethylrhodamine (TAMRA), usually at the 3'-end.
Genome: Wuhan seafood market pneumonia virus isolate 2019-nCoV/USA-AZ1/2020, complete genome Sequence ID: MN997409.1 Length: 29882
Note: A, Adenine; C, Cytosine; G, Guanine; T, Thymine; U, Uracil; R, A or G; Y, C or T; S, G or C; W, A or T; K, G or T; M, A or C; B, C or G or T; D, A or G or T; V, A or C or G; N, any base.
CDC Primer and Probes
|
Name
|
Label 5’
|
Oligonucleotide Sequence (5’>3’)
|
Label 3’
|
Working Conc. [μM]
|
|
|
|
N Protein
|
|
|
|
2019-nCoV_N1-F
|
|
GACCCCAAAATCAGCGAAAT
28287------------28306
|
|
|
|
2019-nCoV_N1-R
|
|
TCTGGTTACTGCCAGTTGAATCTG
28358---------------28335
|
|
20
|
|
2019-nCoV_N1-P
|
FAM
|
ACCCCGCATTACGTTTGGTGGACC
28309---------------28332
|
BHQ1
|
5
|
|
2019-nCoV_N2-F
|
|
TTACAAACATTGGCCGCAAA
29164-------------29183
|
|
20
|
|
2019-nCoV_N2-R
|
|
GCGCGACATTCCGAAGAA
29230------------29213
|
|
20
|
|
2019-nCoV_N2-P
|
FAM
|
ACAATTTGCCCCCAGCGCTTCAG
29188---------------29210
|
BHQ1
|
5
|
|
2019-nCoV_N3-F
|
|
GGGAGCCTTGAATACACCAAAA
28681--------------28702
|
|
20
|
|
2019-nCoV_N3-R
|
|
TGTAGCACGATTGCAGCATTG
28752--------------28732
|
|
20
|
|
2019-nCoV_N3-P
|
FAM
|
AYCACATTGGCACCCGCAATCCTG
28706--------------28727
|
BHQ1
|
5
|
|
Name
|
Label 5’
|
Oligonucleotide Sequence (5’>3’)
|
Label 3’
|
Working Conc. [μM]
|
|
|
|
RNAse Protein
|
|
|
|
RP-F
|
|
AGATTTGGACCTGCGAGCG
|
|
20
|
|
RP-R
|
|
GAGCGGCTGTCTCCACAAGT
|
|
|
|
RP-P RNAse P
|
FAM
|
TTCTGACCTGAAGGCTCTGCGCG
|
BHQ-1
|
5
|
|
Name
|
Label 5’
|
Oligonucleotide Sequence (5’>3’)
|
Label 3’
|
Working Conc. [μM]
|
|
|
|
ORF1ab
|
|
|
|
2019-nCoV-OFP
|
|
CCCTGTGGGTTTTACACTTAA
13342------------13362
|
|
0.6
|
|
2019-nCoV-ORP
|
|
ACGATTGTGCATCAGCTGA
13460------------13442
|
|
0.8
|
|
2019-nCoV-OP
|
FAM
|
CCGTCTGCGGTATGTGGAAAGGTTATGG
13377-------------------13404
|
BBQ1
|
0.1
|
|
|
|
Nucleoprotein- protein N
|
|
|
|
2019-nCoV-NFP
|
|
GGGGAACTTCTCCTGCTAGAAT
28881-------------28902
|
|
0.6
|
|
2019-nCoV-NRP
|
|
CAGACATTTTGCTCTCAAGCTG
28979-------------28958
|
|
0.8
|
|
2019-nCoV-NP
|
FAM
|
TTGCTGCTGCTTGACAGATT
28934-------------28953
|
TAMRA
|
0.1
|
|
|
|
ORF1b-nsp14
|
|
|
|
HKU-ORF1b-nsp14F
|
|
TGGGGYTTTACRGGTAACCT
18778-----------18797
|
|
0.6
|
|
HKU- ORF1b-nsp14R
|
|
AACRCGCTTAACAAAGCACTC
18909-------------18889
|
|
0.8
|
|
HKU-ORF1b-nsp141P
|
FAM
|
TAGTTGTGATGCWATCATGACTAG
18849---------------18872
|
TAMRA
|
0.1
|
|
|
|
Nucleoprotein – Protein N
|
|
|
|
HKU-NF
|
|
TAATCAGACAAGGAACTGATTA
29145--------------29166
|
|
0.6
|
|
HKU-NR
|
|
CGAAGGTGTGACTTCCATG
29254-----------29236
|
|
0.8
|
|
HKU-NP
|
FAM
|
GCAAATTGTGCAATTTGCGG
29196------------29177
|
TAMRA
|
0.1
|
|
Positive Control for insertion into plasmid
|
|
AGTTGACTTCGCAGTGGCTAACTAACATCTTTGGCACTGTTTATGAAAAACTCAAACCCGTCCTTGATTGGCTTG
AAGAGAAGTTTAAGGAAGGTGTAGAGACCCTGTGGGTTTTACACTTAAAAACACAGTCTGTACCGTCTGCGGTAT
GTGGAAAGGTTATGGCTGTAGTTGTGATCAACTCCGCGAACCCATGCTTCAGTCAGCTGATGCACAATCGTTTTT
ACTCCAGGCAGCAGTAGGGGAACTTCTCCTGCTAGAATGGCTGGCAATGGCGGTGATGCTGCT
|
|
Name
|
Label 5’
|
Oligonucleotide Sequence (5’>3’)
|
Label 3’
|
Working Conc. [μM]
|
|
|
|
RNA-dependent RNA polymerase
|
|
|
|
RdRP_SARSr-F2
|
|
GTGARATGGTCATGTGTGGCGG
15431-------------15452
2019-nCoV genome
|
|
0.6
|
|
RdRP_SARSr-R1
|
|
CARATGTTAAASACACTATTAGCATA
15460-----------------15435
Coronavirus BtRs-BetaCoV/YN2018D, complete genome. Sequence ID: MK211378.1 Length: 30213
|
|
0.8
|
|
RdRP_SARSr-P1
|
FAM
|
CCAGGTGGWACRTCATCMGGTGATGC
145---------------------170
Bat coronavirus isolate B18-171 RNA-dependent RNA polymerase (RdRp) gene, partial cds. Sequence ID: MK991947.1 Length: 387
|
BBQ1
|
0.1
|
|
RdRP_SARSr-P2
|
FAM
|
CAGGTGGAACCTCATCAGGAGATGC
15470----------------15494
Specific for 2019-nCoV, will not detect SARSCoV.
Wuhan seafood market pneumonia virus isolate 2019-nCoV/USA-AZ1/2020, complete genome Sequence ID: MN997409.1Length: 29882
|
BBQ1
|
0.1
|
|
|
|
E gene
|
|
|
|
E_Sarbeco_F1
|
|
ACAGGTACGTTAATAGTTAATAGCGT
26269------------------26294
|
|
0.4
|
|
E_Sarbeco_R2
|
|
ATATTGCAGCAGTACGCACACA
26381---------------26360
|
|
0.4
|
|
E_Sarbeco_P1
|
FAM
|
ACACTAGCCATCCTTACTGCGCTTCG
26332------------------26357
Sequence ID: MN997409.1. Also detects MK211378.1, MK062184.1 etc.
|
BBQ
|
0.2
|
|
Name
|
Label 5’
|
Oligonucleotide Sequence (5’>3’)
|
Label 3’
|
Working Conc. [μM]
|
|
|
|
nCoV Envelope Gene
|
|
|
|
CoV-EFP
|
|
ACTTCTTTTTCTTGCTTTCGTGGT
26295 ----------------26318
|
|
20
|
|
CoV-ERP
|
|
GCAGCAGTACGCACACAATC
26376------------26357
|
|
20
|
|
CoV-EP
|
FAM
|
CTAGTTACACTAGCCATCCTTACTGC
26326----------------26351
|
BBQ1
|
5
|
|
|
|
ORF1b-nsp14
|
|
|
|
CoV-EFP
|
|
ACTTCTTTTTCTTGCTTTCGTGGT
26295 ----------------26318
|
|
0.6
|
|
CoV-ERP
|
|
GCAGCAGTACGCACACAATC
26376------------26357
|
|
0.8
|
|
CoV-EP
|
FAM
|
CTAGTTACACTAGCCATCCTTACTGC
26326----------------26351
|
BBQ1
|
0.1
|
|
|
|
N Protein
|
|
|
|
CoV-EFP
|
|
TAATCAGACAAGGAACTGATTA
29145---------------29166
|
|
0.6
|
|
CoV-ERP
|
|
TAATCAGACAAGGAACTGATTA
29145---------------29166
|
|
0.8
|
|
CoV-EP
|
FAM
|
GCAAATTGTGCAATTTGCGG
29196--------------29177
|
BBQ1
|
0.1
|
Reference
https://www.who.int/docs/default-source/coronaviruse/peiris-protocol-16-1-20.pdf?sfvrsn=af1aac73_4
Liu G, Li H, Zhao S, Lu R, Niu P, Tan W. Viral and Bacterial Etiology of Acute Febrile Respiratory Syndrome among Patients in Qinghai, China.Biomed Environ Sci. 2019 Jun;32(6):438-445. https://www.sciencedirect.com/science/article/pii/S0895398819301230
Huang et al. 2020: https://www.sciencedirect.com/science/article/pii/S0140673620301835
---...----