Specific DNA probes allow the detection and discrimination of specific nucleic acid target sequences. In the early years of probe development, DNA probe development utilized material from natural sources or cloning DNA. However, synthetic DNA hybridization probes are now more commonly used for diagnostics.
Extensive research showed that using specific oligonucleotides allows sensitive detection and distinguishing of various virus types, including Hepatitis B virus, HSV type 1 and HSV type 2 virus, HIV viruses, and coronaviruses such as SARS-CoV and SARS-CoV-2 in clinical isolates.
Also, specific modification of synthetic oligonucleotides enables the design and synthesis of non-radioactive in situ hybridization probes. Biotin-labeled probes are one example. Over three-fourths of men infected with human immunodeficiency virus (HIV) also have at least one herpes virus detected in their semen. In this case, the cytomegalovirus (CMV) is most prevalent. The presence of CMV is associated with higher T-cell immune activation and with HIV disease progression in treated and untreated individuals.
HIV is known to have a significant genetic variability; therefore, achieving accurate quantification by real-time PCR may require primers and probes specific for each viral variant. An HIV database is available to download sequences for particular variants.
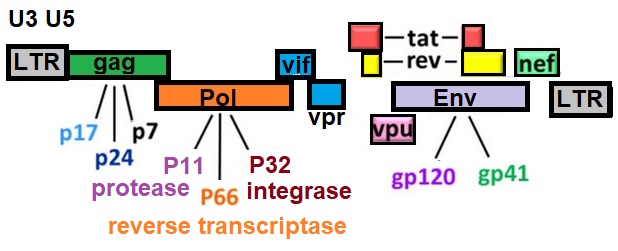
HIV Genome
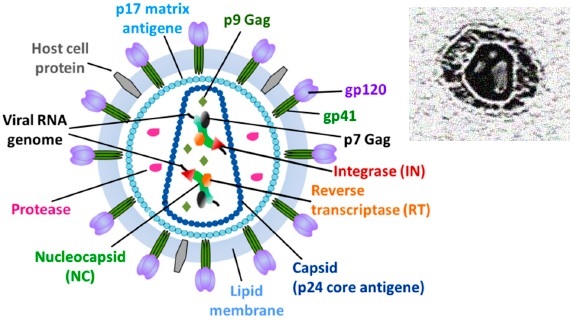
Structural Model of the HIV Particle
All primer and probes can be biotinylated or labeled as needed, for example with fluorophores to create hybridization probes.
Table 1: Primer and Probes for HIV-1 Diagnostics using RT-PCR
|
Primer
|
Sequence
|
|
|
Early Region
|
|
Cr1
|
TCTCTGGCTAACTAGGGAACCCACTGCTT
|
|
Cr2
|
TGACTAAAAGGGTCTGAGGGATCTCTAGTTACCAG
|
|
|
gag region
|
|
ts5’gag
|
CAAGCAGCCATGCAAATGTTAAAAGA
|
|
skcc
|
TACTAGTAGTTCCTGCTATGTCACTTCC
|
|
SK38
|
ATAATCCACCTATCCCAGTAGGAGAAAT
|
|
SK39
|
TTTGGTCCTTGTCTTATGTCCAGAATGC
|
|
|
pol region
|
|
mf209
|
AAAGCGTCTAGCCATGGCGTTAGTA
|
|
mf302
|
CAAATTTCTACTAATGCTTTTATTTTTTC
|
|
|
tat region
|
|
mf1
|
CTTAGGCATCTCCTATGGCAGGAA
|
|
mf238
|
GCTATTATTGCTGCTACTACTAATGCTACTA
|
|
|
sa7 region
|
|
mf222
|
GGCAGGGATATTCACCATTATCGTTTCAGA
|
|
mf83
|
GGATCTGTCTCTGTCTCTCTCTCCACC
|
|
|
nef region
|
|
mf345
|
AATCAGGGAAGTAGCCTTGTGT
|
|
mf346
|
GAGGTGGGTTTTCCAGT
|
|
Beacon
|
5′-FAM-CGGGAGTACTCACCAGTCGCCGCCCCTCGCCCTCCCG-DABCYL-3′
|
|
HIV-1 (SK-19) Probe
|
ATCCTGGGATIAAATAAAATAGTAAGAATGTATAGCCCTAC
|
.
Reference
Althaus CF, Gianella S, Rieder P, et al. Rational design of HIV-1 fluorescent hydrolysis probes considering phylogenetic variation and probe performance. J Virol Methods. 2010;165:151–60. [Pdf]
Christopherson, C., Kidane, Y., Conway, B., Krowka, J., Sheppard, H., Kwok, S., 2000. PCR-Based assay to quantify human immunodeficiency virus type 1 DNA in peripheral blood mononuclear cells. J.Clin.Microbiol. 38, 630-634. [PMC]
Fischer, M., Joos, B., Hirschel, B., Bleiber, G., Weber, R., Günthard, H.F., 2004. Cellular viral rebound after cessation of potent antiretroviral therapy predicted by levels of multiply spliced HIV-1 RNA encoding nef. J Infect Dis 190, 1979-88. [Pdf]
HIV database
Kaiser, P., Joos, B., Niederoest, B., Weber, R., Günthard, H.F., Fischer, M., Study, T.S.H.C., 2007. Productive Human Immunodeficiency Virus 1 Infection in peripheral Blood Predominantely Takes Place in CD4/CD8 double negative T Lymphocytes. J Virol 81, 9693-9706. [PMC]
Lewin, S.R., Vesanen, M., Kostrikis, L., Hurley, A., Duran, M., Zhang, L., Ho, D.D., Markowitz, M., 1999. Use of Real-Time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J.Virol. 73, 6099-6103. [PMC]
Musumeci D, Riccardi C, Montesarchio D. G-Quadruplex Forming Oligonucleotides as Anti-HIV Agents. Molecules. 2015 Sep 22;20(9):17511-32. [PMC]
---...---