The innate immune system discriminates between self and non-self mainly based on receptors recognizing non-self molecules present in pathogens, but not present in the host. Recent observations showed that inosine present in the epitranscriptome and the adenosine deaminases acting on RNA (ADAR1) protein establishes innate immune tolerance for endogenous host double-stranded RNA. So far over a hundred (100) million adenosine to inosine editing sites have been identified in the human transcriptome.
The term “transcriptome” refers to the sum of all the messenger RNA molecules expressed from the genes of an organism. However, the term “epitranscriptome” refers to all biochemical modifications of RNA within a cell.
ADARs are editing enzymes converting adenosine (A) to inosine (I) in duplex RNA. This modification has wide-ranging consequences on RNA function.
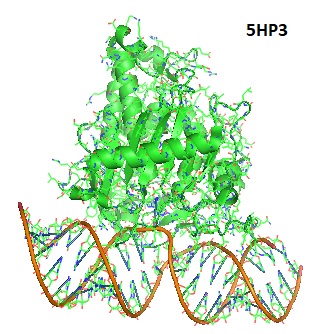
Structure of hADAR2d E488Q bound to the Bdf2-C RNA duplex. Matthews et al. 2016. Matthews et al. in 2016 reported four crystal structures of the human ADAR2 deaminase domain bound to RNA duplexes bearing a mimic of the deamination reaction intermediate. These structures help explaining the basis of the ADAR deaminase domain's dsRNA specificity, its base-flipping mechanism, and its nearest-neighbor preferences. Studying these structures may help to increase our understanding of the effects of ADAR mutations associated with human disease.
Inosine sites are mostly found in embedded Alu sequences which form potentially innate immune-stimulating dsRNA hairpins in transcripts. The nucleotide inosine is commonly found in tRNAs and is formed when hypoxanthine attaches to a ribose ring via a β-N9-glycoside bond.
Viral nucleic acids are detected by innate immune sensors that act as readers of epitranscriptome RNA modifications. 'A-to-I editing' can alter a protein's coding sequence. In humans, this is crucial for keeping the innate immune response in check.
Inosine can pair with any natural base allowing the design of oligonucleotide primers spanning target sequences containing single nucleotide polymorphisms (SNPs). The use of inosine-containing amplification primers allows the amplification of specific DNA fragments from different alleles.
Because inosine pairs preferentially with cytidine (C) its introduction to RNA via the action of ADARs destabilizes double-stranded RNA by changing AU base-pairs to IU mismatches.
Conclusion
• The innate immune system discriminates between self and non-self.
• The discrimination is based on receptors recognizing non-self-molecules present in pathogens, but not present in the host.
• Viral nucleic acids are detected by innate immune sensors that act as readers of epitranscriptome RNA modifications.
• Inosine present in the epitranscriptome and the adenosine deaminases acting on RNA (ADAR1) protein establishes
innate immune tolerance for endogenous host double-stranded RNA.
• Inosine sites are mostly found in embedded Alu sequences which form potentially innate immune-stimulating dsRNA
hairpins in transcripts.
• Inosine pairs preferentially with cytidine (C) and when present in RNA destabilizes double-stranded RNA by changing
AU base-pairs to IU mismatches.
• Inosine-containing amplification primers allows amplification of specific DNA fragments resulting from different alleles.
Reference
A-to-I editing
ADAR2 structure 5HP3
Bass BL, Weintraub H.; An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988 Dec 23;55(6):1089-98.
[PubMed]
Ben-Dov E (2006). "Advantage of Using Inosine at the 3′ Termini of 16S rRNA Gene Universal Primers for the Study of Microbial Diversity". Applied and Environmental Microbiology. 72: 6902-6906. [P
MC]
Epitrancriptome and Innate Immunity
George CX, Gan Z, Liu Y, Samuel CE. Adenosine deaminases acting on RNA, RNA editing, and interferon action. J Interferon Cytokine Res. 2011;31(1):99-117. [
PMC]
Kim, D. D., Kim, T. T., Walsh, T., Kobayashi, Y., Matise, T. C., Buyske, S., & Gabriel, A. (2004). Widespread RNA editing of embedded alu elements in the human transcriptome. Genome research, 14(9), 1719-25. [
PMC]
Köstler K, Rosemeyer H. Towards the synthesis of inosine building blocks for the preparation of oligonucleotides with hydrophobic alkyl chains between the nucleotide units. Molecules. 2009;14(11):4326-36. [
PMC]
Erez Y Levanon, Eli Eisenberg, Rodrigo Yelin, Sergey Nemzer, Martina Hallegger, Ronen Shemesh, Zipora Y Fligelman, Avi Shoshan, Sarah R Pollock, Dan Sztybel, Moshe Olshansky, Gideon Rechavi & Michael F Jantsch; Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nature Biotechnology volume 22, pages1001–1005 (2004). [
Link]
Matthews MM, Thomas JM, Zheng Y, et al. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat Struct Mol Biol. 2016;23(5):426-33. [
PMC]
Nucleic Acid Sequence Wiki
The epitranscriptome Collection
---...---