Selective 2’-Hydroxyl Acylation and Primer Extension or SHAPE
The majority of RNA molecules only appear to function when they exist in the correct secondary or tertiary structure by folding back onto themselves into difficult to predict three-dimensional structures. Many RNAs have specific regions with local structural flexibility or can transition from one conformation to another. Known RNA motif families are GNAA tetraloop, kink-turn, sarcin-ricin, and T-loop, but other potential motifs are also present in ribosomal RNA, sgRNA, SRP RNA, riboswitch and ribozyme.
To understand the function of RNAs, accurate, and comprehensive knowledge of the base-paired secondary structure and the identification of nucleotides involved in tertiary structure is needed. Known functional RNA components with three-dimensional (3D) conformation fall under the category of RNA motifs.
With that in mind, in years past, researchers have used a veriety of chemical and enzymatic reagents for mapping RNA secondary structures. However, many of these approaches suffer from shortcomings such as:
-
Low discriminative power: For many reagents, the magnitude of discrimination between single-stranded and base-paired regions can be small, and helix termini and some base pairs are often reactive.
-
Infrequent results: The obtained information is often of limited value since a given reagent may react with only a subset of the four RNA nucleotides or backbone sites.
-
Low selectivity: Therefore, multiple reagents are often employed for the interrogation of all positions in an RNA molecule
-
No unifying rule sets: Historically, it has been challenging to develop unifying rule sets for classes of structures that react versus that do not.
In 2005, Merino et al. reported an alternative approach for mapping RNA structures using the observation that the chemical reactivity of the 2’-ribose position is influenced by the adjacent 3’-phosphodiester anion.
Earlier in 1999, Chamberlin and Weeks started evaluating the chemical selectivity of 2′-amine acylation using a model oligonucleotide substrate containing either all ribose nucleotides or a single 2′-amino-cytidyl substitution. Chamberlin and Weeks investigated reagents allowing selective acylation of the 2'-ribose position for their use in mapping RNA structures. Also, the magnesium ion-dependent conformational changes in tRNAAsp transcripts containing single 2′-amine substitutions per transcript were studied as well. Chamberlin and Weeks found that acylation of synthetic 2’-amine-substituted nucleotides forming the 2’-amide product is strongly confined by the underlying local flexibility of the nucleotide.
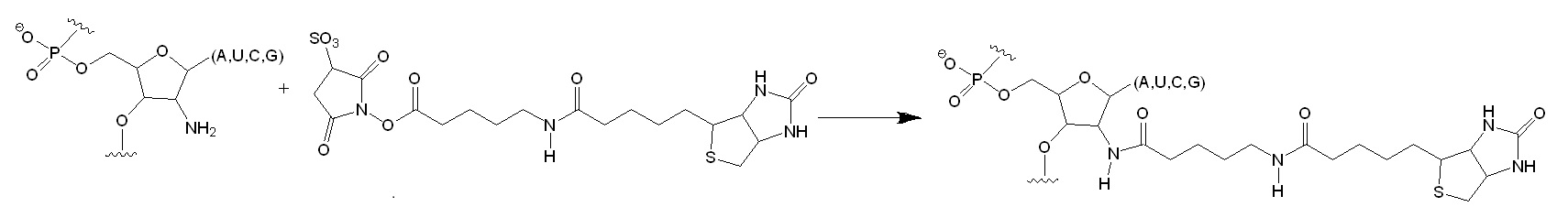
Figure 1: Selective Acylation of 2′-Amine Positions in RNA. The oligonucleotide substrate was treated with a succinimidyl ester. The incorporation of the biotinyl group was monitored as a slowly migrating band in a denaturing polyacrylamide gel. The all-ribose substrate reacted very slowly with the reagent (4% conversion in 60 min) as compared to the 2′-amine substituted oligonucleotide (97% conversion in 60 min). The slow modification of the all-ribose substrate is attributed to the reaction of base arylamines with the succinimidyl ester.
The researchers found that under denaturing conditions, all 2′-amine substituted RNA positions show similar reactivity. However, when tRNAAsp transcripts are refolded under native conditions (10 mM Mg2+, 100 mM NaCl), positions involved in base pairing and known tertiary interactions, including base triples and loop-loop interactions, are protected from modification. When no magnesium ions are present, the acceptor, T- and anticodon stems forms stable helices leading to a relatively low 2′-amine reactivity. These results suggested an interdependence between the formation of the D-stem helix and tertiary structure folding for yeast tRNAAsp transcripts. The researchers used this chemical approach for mapping local RNA flexibilities. Reported experimental results were consistent with prior biophysical and biochemical studies showing its utility for mapping local nucleotide environments on small amounts of RNA molecules.
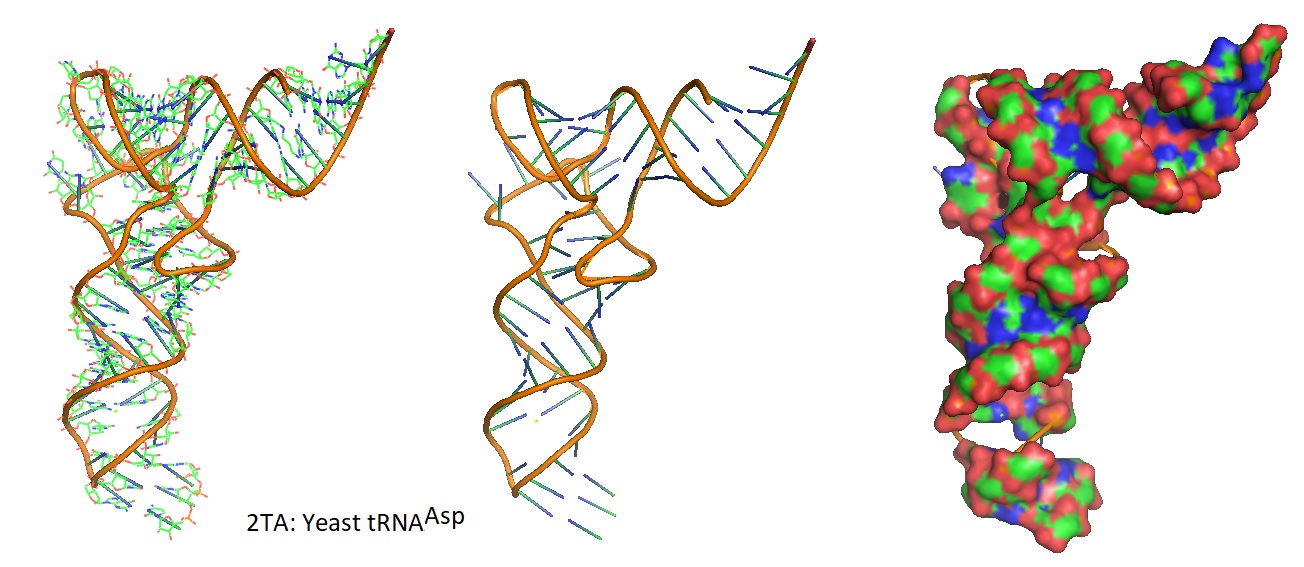
Figure 2: Models of the 3D structure of yeast tRNAAsp from Asp and Phe transfer RNA crystals 2TA (Westhof et al. 1988).
Mortimer and Weeks set out to find a more selective fast-acting reagent needed for accurate analysis of RNA secondary and tertiary structures using SHAPE chemistry. The reagent selected needs to allow chemically assisted RNA structure analysis with the following criteria:
- Enable a chemically straightforward approach,
- Use of a single reagent reactive to all four nucleotides,
- Is self-quenching,
- Has a short reaction time, and
- Yields accurate results with complex RNA molecules of known structure.
After testing the reagent N-methylisatoic anhydride (NMIA) for is use in SHAPE the scientists decided to develop a faster reacting reagent. The reagent 1-methyl-7-nitroisatoic anhydride (1M7) appears to fulfill these criteria.
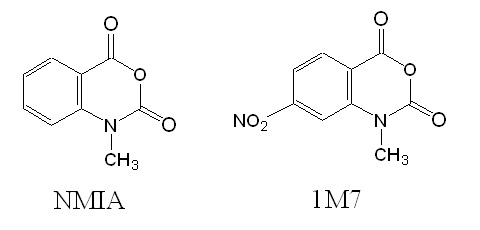
Figure 3: Structures of reagents NMIA and 1M7.
In 1M7, the para nitro substituent is strongly electron-withdrawing leading to an increase in adduct formation as well as hydrolysis rates. Experimental verification allowed the researcher to conclude that 1M7 has the correct chemical characteristics for a fast-acting and self-quenching reagent for its use in SHAPE chemistry.
A SHAPE experiment performed on the RNase P domain under condition that stabilized the native tertiary fold allowed identifying 2’-O-adduct formation as stops of primer extension, using fluorescently labeled DNA primers, as resolved by capillary electrophoresis. Experiments were performed in the presence and absence of Mg2+ ions to test how magnesium ions influence the flexibility of the structure.
To conclude, SHAPE chemistry performed with 1M7 allowed the researchers to generate an accurate report of the known structure of the RNase P specificity domain under native conditions.
Reference
1988: Westhof, Dumas, and Moras; Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals. Acta Crystallogr A 1988 Mar 1;44 (Pt 2): 112-123.
2000: Chamberlin and Weeks; Mapping local nucleotide flexibility by selective acylation of 2-amine substituted RNA. J. Am. Chem. Soc. 2000, 122, 216-224.
2005: Edward J. Merino, Kevin A. Wilkinson, Jennifer L. Coughlan, Kevin M. Weeks; RNA Structure Analysis at Single Nucleotide Resolution by Selective 2‘-Hydroxyl Acylation and Primer Extension (SHAPE). J. Am. Chem. Soc. 2005, 127, 12, 4223-4231. https://doi.org/10.1021/ja043822v.
2007: Stefanie A. Mortimer and and Kevin M. Weeks; A Fast-Acting Reagent for Accurate Analysis of RNA Secondary and Tertiary Structure by SHAPE Chemistry. Journal of the American Chemical Society 2007 129 (14), 4144-4145. DOI: 10.1021/ja0704028. https://pubs.acs.org/doi/full/10.1021/ja0704028.
2018: RNA motifs: Ping Ge, Shahidul Islam, Cuncong Zhong, Shaojie Zhang, De novo discovery of structural motifs in RNA 3D structures through clustering, Nucleic Acids Research, Volume 46, Issue 9, 18 May 2018, Pages 4783–4793, https://doi.org/10.1093/nar/gky139.
---…----