Numbering convention for nucleotides
Nucleic acids are the building blocks for polymers of nucleotides. Oligonucleotides contain, store and transmit instructions about proteins and quantities of these a cell needs to function. This information system is called the genetic code. The chemical structures that contain and decode the code are called deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). The DNA molecule stores the information and the active agent RNA decodes the stored information. Both DNA and RNA are linear polymers in their primary structures composed of monomers or single chemical units or building blocks called nucleotides. Cellular RNA molecules can range in length from leas than one hundred to thousands of monomers. However, the number of monomers in DNA can exceed a hundred million.
DNA and RNA are made up of only four different nucleotides. A nucleotide has three characteristic parts: (A) A nitrogenous or nitrogen-containing organic base, (B) a pentose or five-carbon sugar molecule, and (C) a phosphate group. Bases and pentoses are heterocyclic compounds. The pentose in RNA is always a ribose, however, in DNA it is a deoxyribose. In addition, one of the four nucleic acid bases differs between the two polymers.
Adenine, guanine, and cytosine are present in both, DNA and RNA. Thymine is present in DNA, and uracil is only present in RNA. Abbreviations of these bases are A, G, C, T, and U, respectively. Components of the bases are either purines (A and G) or pyrimidines (C, T, or U). Figure 1 shows the general structure of nucleotides illustrating the numbering convention for the pentose ring. The ribose structure is shown in the Haworth projection. Phosphate groups join the nucleic acid monomers together in a linear manner. Phosphate groups are attached to the 3’ and 5’ positions of the ribose sugars. Therefore, the repeating nucleic acid unit is a 3’,5’-nucleotide. The two classes of bases are often abbreviated a Y, originating from pyrimidine, and R, originating from purine. The short syllable for the phosphate group is P.
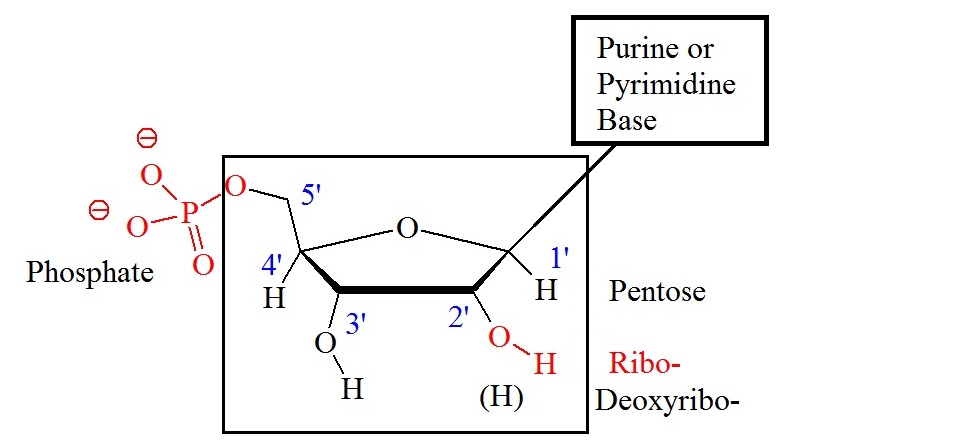
Figure 1: General structure showing the numbering convention for the pentose ring. The carbon atoms of the pentoses are numbered with primes.
.jpg)
Figure 2: Ribonucleic acid (RNA) fragment containing adenosine (A), guanosine (G), uridine (U), cytidine (C) linked by 3',5'-phosphodiester bonds. The chain direction from 5' to 3' is indicated by an arrow. The atom numbering scheme is indicated in the nucleotide units.
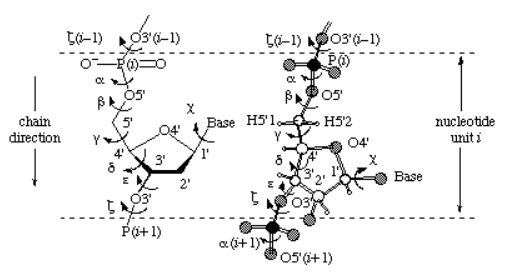
Figure 3: Atomic numbering scheme and definitions. Torsion angles for an oligonucleotide chain are shown as well.
.jpg)
Figure 4: DNA and RNA building blocks. The structures and their molecular weights that make up the building block for oligonucleotides are shown.
Small concentrations of nucleosides are present in cells and extracellular fluids. The naming of the four ribonucleosides and deoxyribonucleosides is shown in table 1. Nucleotides that have phosphate groups attached are referred to as nucleoside phosphates.
Table 1: Naming Nucleosides and Nucleotides
Reference
IUPAC nomenclature: http://www.chem.qmul.ac.uk/iupac/misc/naabb.html
Books to review:
Genomes, 2nd edition. Terence A Brown. Department of Biomolecular Sciences, UMIST, Manchester, UK. Oxford: Wiley-Liss; 2002. ISBN-10: 0-471-25046-5
Molecular Biology of the Cell. 4th edition. Alberts B, Johnson A, Lewis J, et al. New York: Garland. Science; 2002.
Molecular Cell Biology. 4th edition. Lodish H, Berk A, Zipursky SL, et al. New York: W. H. Freeman; 2000.
The Cell: A Molecular Approach. 2nd edition. Cooper GM. Sunderland (MA): Sinauer Associates; 2000.
Lehninger Principles of Biochemistry and Absolute Ultimate Guide Cox, Michael M., Nelson, David L., Lehninger, Albert L. ISBN-13: 9781429212410. ISBN: 1429212411 Edition: 5 Pub Date: 2008. Publisher: W. H. Freeman Company.
Principles of Nucleic Acid Structure (Springer Advanced Texts in Chemistry) Paperback – October 19, 1988 Wolfram Saenger.
Principles of Nucleic Acid Structure. Stephen Neidle. ISBN: 978-0-12-369507-9.
