What are splice-switching oligonucleotides (SSOs)?
Splice-switching oligonucleotides are short, synthetic, oligonucleotides that act as antisense oligonucleotides. Oligonucleotides containing modified nucleic acids, for example bridged nucleic acids (BNAs), base-pair with pre-mRNA with high affinity and disrupt normal splicing of transcripts by blocking the RNA–RNA base-pairing or protein–RNA binding interactions that occur between components of the splicing machinery and pre-mRNAs. Splice-switching antisense oligonucleotides containing a backbone modified with 2’-O-methyl-phosphorothioate groups and approximately 60% BNAs, as well as two BNA-modified nucleotides at the 3’-end and one BNA-modified nucleotide at the 5’-end appear to work well for controlling and modulating the expression of specific exons hence they can act as antisense oligonucleotides enabling modulation and regulation of splicing events.
Exon skipping oligonucleotides can be designed rationally. Design approaches involve the selection of an antisense sequence targeting the exon that is supposed to be skipped. Design parameters to be taken into account include the activity of the splice-switching oligonucleotide, the melting temperature, the guanine-cytosine content, the length of the oligonucleotide, as well as the secondary structure or sequence motif corresponding to a splicing signal of the target RNA that influence the activity of the splice-switching oligonucleotide. Since the synthesis of modified oligonucleotides is now routinely done in an automated fashion, splice-switching oligonucleotides, containing modified or unmodified nucleic acid, are now commercially available.
What is splicing?
According to the NCI Dictionary of Genetics Terms “splicing” is defined as the “process by which introns, the noncoding regions of genes, are excised out of the primary messenger RNA transcript, and the exons (i.e., coding regions) are joined together to generate mature messenger RNA. Mature mRNA then serves as the template for synthesis of a specific protein.” Proper expression of most protein-coding genes requires the splicing of pre-mRNA. In other words, splicing refers to the editing of newly transcribed pre-messenger RNA (pre-mRNA), the removal of introns and the joining or ligation of exons. Genes encoded in the nucleus are spliced within the nucleus either during or after transcription.
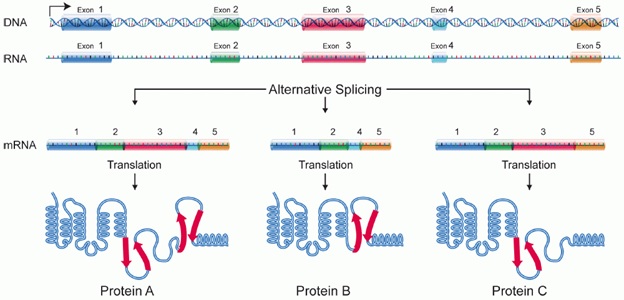
Illustration explaining the process of “Alternative, or Differential Splicing.” As a result of splicing a single gene can code for multiple proteins. During alternative splicing a specific exons of a gene may be included within or excluded from the final messenger RNA (mRNA) produced from that gene.
Reference
Alternative splicing
Havens MA, Hastings ML. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016;44(14):6549-63.
Ryszard Kole, Tiffany Williams and Lisa Cohen; RNA modulation, repair and remodeling by splice switching oligonucleotides Acta Biochemica Polonica. Vol. 51 No. 2/2004 373–378.
NCI Dictionary of Genetics Terms splicing
Pires VB, Simões R, Mamchaoui K, Carvalho C, Carmo-Fonseca M. Short (16-mer) locked nucleic acid splice-switching oligonucleotides restore dystrophin production in Duchenne Muscular Dystrophy myotubes. PLoS One. 2017;12(7):e0181065. Published 2017 Jul 24. doi:10.1371/journal.pone.0181065.
---...---