Why do we need cell line identification and authentication?
As reported in the literature, cell lines are frequently contaminated or misidentified. A high percentage of cell lines are cross-contaminated with cells from other cell lines. This phenomenon is known as “cell line cross-contamination” or (CLCC). According to MacLeod, CLCC occurs primarily during the establishment of a cell culture. Presumably, new cell cultures are contaminated during the early phases of their establishment. Cell line cross-contamination is a result of continuous cell line culture. To avoid misidentification quality controls need to be carried out to address this issue.
Sometimes, cross-contaminating cell lines can overgrow the original cell line. Cross-contamination has caused the number of misidentified circulating cell lines to be unacceptably high. In particular, the use of cancer cell line models may lead to wrong results if not identified or authenticated correctly. Therefore scientists have recommended that researchers should provide authenticities of all experimental cell lines used in experiments before publishing experimental results.
The reported misidentification of cell lines is estimated to be between 16 % to 36 %. These findings are based on the analysis of submitted cell lines. One of the best examples is the false description of a HeLa cervical cancer cell line. As a result, cell line identification has now become an essential method for the identification of human cell lines used in research. Cell lines are important reagents for experimental biomedical sciences. A large number of cell lines have been derived from patients. Among them are cell lines from patients with multiple myelomas, plasma cell leukemia, Plasmacytoma and many others.
Bio-Synthesis Inc. offers Custom Cell Line Identification
Cell lines can be tested and identified using multiallelic variable number of tandem repeats (VNTR). HLA typing and DNA fingerprinting using short tandem repeat (STR) and a variable number of tandem repeats for intra-species cross-contamination have been used for cell line identification. However, DNA fingerprinting hase become the method of choice for cell line identification.
Cell banks that archive and distribute cell lines are responsible for guaranteeing the authenticity of cell lines and will need to ensure cell line authenticity via DNA finger printing.
As pointed out by John M. Butler, STR typing enables the rapid discovery of cross-contamination between cell lines. Since STR typing is now used as the major tool for characterization of human cell lines this approach has also been called “cell culture forensics.” The American Type Culture Collection (ATCC), the National Institutes of Biomedical Innovation containing the Japanese Collection of Research Bioresources (JCRB) Cell Bank, the “Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH” (DSMZ), as well as others, offer cell lines for research and have created databases containing information for many cell lines.
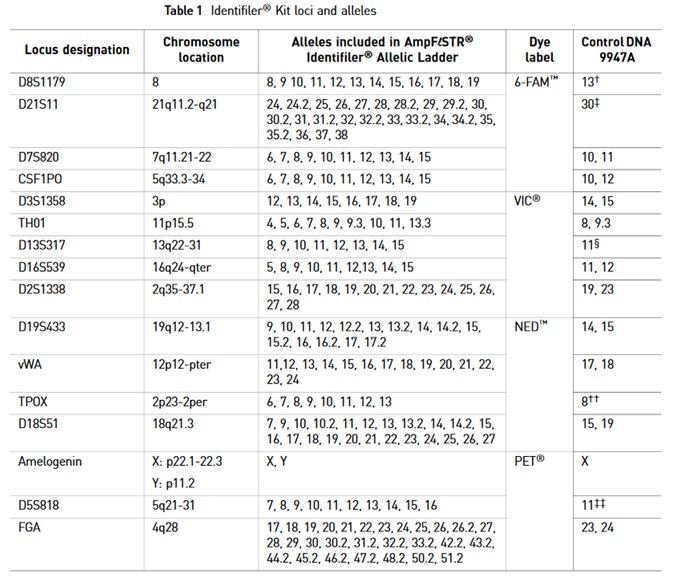
PCR-based identification kits use short tandem repeat (STR) amplifications assays. For example, the AmpFιSTR®PCR Amplification Kit is a short STR multiplex assay that amplifies 15 tetranucleotide repeat loci and the Amelogenin gender-determining marker in a single PCR amplification experiment. Fluorescent multi-color dye technology allows the analysis of multiple loci. Alleles with overlapping size ranges are also included. Locus-specific primers labeled with different colored dyes allow distinguishing overlapping loci.
STR markers are polymorphic DNA loci containing repeated nucleotide sequences. STR repeat units can range from two to seven nucleotides in length. However, the number of nucleotides per repeat unit is the same for a majority of repeats within an STR locus, but the number of repeat units at any STR locus may differ. Therefore alleles of many different lengths are possible. Polymorphic STR loci are very useful for human identifications as well as for cell line identification.
STR loci can be amplified using the polymerase chain reaction (PCR) process. PCR products are analyzed by electrophoresis to separate alleles according to size. Methods such as fluorescent dye labeling, silver staining, or fluorescent dye staining can be used for detection.
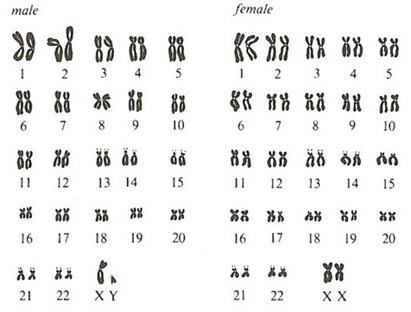
The human genome contains approximately three (3) billion base pairs in a single copy. Because of the Human Genome Project, a reference genome is now available.
Genomic DNA found in the nucleus of a human’s cell is divided into chromosomes. In chromosomes, DNA is densely packaged around proteins called histones. Twenty-two (22) matching pairs of autosomal chromosomes and two (2) sex chromosomes make up the human genome. Hence, the cell of normal humans contains 46 different chromosomes. Females are designated XX and males are designated XY, because they contain a single copy of an X and Y chromosome. Somatic chromosomes in cells in the human body are in a diploid state. They contain two sets of chromosomes. However, gametes (sperms or eggs) are in a haploid state. The combination of an egg cell with a sperm cell results again in a diploid state. DNA in chromosomes is divided into “coding” and “non-coding” sequence regions. Coding regions are known as “genes” and can contain a few thousand to tens of thousands base pairs. Since the finished Human Genome Project, we now know that the human genome codes for ~30,000 protein-coding genes. Therefore genes make up ~5% of human genomic DNA.
Human polymorphic markers are found in specific loci!
Markers useful for human identity or cell line identification testing are found in “non-coding” regions, either between genes or within genes (i.e. introns).
Polymorphic or variable markers are found throughout the non-coding regions of the human genome. The location or position of a gene or DNA marker is referred to as a “locus” (plural: loci). Thousands of these loci have now been mapped to specific regions of human chromosomes.
Pairs of chromosomes are called “homologous” since they are the same size and contain the same genetic structure. One chromosome in each pair is inherited from the mother and one from the father. Therefore the DNA in the two homologous chromosomes may not be identical since mutations could have occurred over time.
Alternative regions in homologous chromosomes are called “alleles.”
If DNA sequences in two alleles at a genetic locus on homologous chromosomes differ they are called “heterozygous,” but if they are similar they are called “homozygous.”
A ‘genotype” refers to alleles that are present at a genetic locus. If two alleles are present at one locus, AA and aa, three genotypes are possible, AA, Aa, and aa. A “DNA profile” is the combination of genotypes obtained for multiple loci.
DNA profiling refers to the process of determining the genotype present at specific locations along the DNA molecule.
A Brief Nomenclature for DNA Markers
Commonly used STR markers are found in sequence repeats present in satellites (~100 to 1000 repeated bases), minisatellites (VNTR = variable number tandem repeat; ~10 to 100 repeated bases), microsatellites (STR = short tandem repeat; 2 to 6 repeated bases. Most commonly used in forensics).
Examples:
STR markers within a gene: ( HUM)THO1
HUM = human; TH = thyrosine hydroxylase gene located on chromosome 11; 01= repeat region is located within intron 1.
STR markers outside gene regions: D16S539
D = DNA; 16 = chromosome; S = single copy sequence; 539 = 539th locus described on chromosome 16.
The vast majority of DNA molecules, approximately over 99.7%, are the same between humans. Only a small fraction, approximately 0.3% or close to 10 million nucleotides, differs between individual people. These variable regions provide the information for human or cell line identification tests.
Two forms of variation are possible at the DNA level (Butler, 2005):
1. Sequence Polymorphisms, and
2. Length polymorphisms.
Example 1: Sequence polymorphism
-----AGACTAGACATT----
-----AGATTAGGCATT----
Example 2: Length polymorphism
----(AATG)(AATG)(AATG)---- 3 repeats
----(AATG)(AATG)---- 2 repeats
Example of the DNA sequence in a STR repeat region
---->
1 2 3 4 5 6
5’-TTTCCC TCAT TCAT TCAT TCAT TCAT TCAT TCACCATGGA-3’
3’-AAAGGG AGTA AGTA AGTA AGTA AGTA AGTA AGTGGTACCT-5’
6 5 4 3 2 1
<----
Using the top strand versus using the bottom strand results in different repeat motifs and starting positions. TCAT repeat units are found in the top strand and TGAA repeat units are found in the bottom strand. According to guidelines issued by the International Society of Forensic Genetics (ISFH; http://www.isfg.org) the top strand should be used for the design of STR markers using alleles.
For STR based typing usually length polymorphism based markers are used. As a rule of thumb: The more multiple markers are used for testing the greater the chance that two unrelated individuals will have different genotypes.
PCR base STR analysis
PCR-based STR analysis has become the method of choice for STR analysis because the small size of STR loci improves the chance of obtaining a good result, especially when small amounts of DNA or degraded DNA are in the sample. Because of the small size range of STR loci, co-amplification can be achieved, and typing is possible from a single PCR. Discrete sizes of STR alleles allow for easier interpretation of results. PCR-based tests are usually rapid, generating results within 24 hours, and lend themselves to automatization and standardization. Automation and standardization ensures that testing results in reproducible results.
A typical protocol for cell line identification
1. DNA is isolated from the cell pellets, tissues or cryovial (frozen cells) with
a DNA extraction kit and purified with a Qiagen or Zymo DNA extraction kit.
2. Quantitation of the extracts can be performed using a
NanoDropTM spectrophotometer.
3. Usually, DNA amplification and fragment fluorescent labeling is
performed using the PCR System thermal cycling instrument and
the IdentifilerTM AmpF/STR® PCR kit.
4. Approximately 20.0 ng/µl of purified DNA is used for amplification.
5. Capillary electrophoresis and fragment detection are performed
with a 310 Genetic Analyzer with Data Collection software v 3.0
(Applied Biosystems).
6. GeneMapper ID® (Applied Biosystems) software is used to assign fragment
repeat numbers which becomes collectively the STR type for the individual
cell line identified.
The AmpFι STR®PCR Identifiler Kit uses the following STR marker for genotyping in a single reaction:
A brief glossary of STR testing
|
Humans = Homo Sapiens
|
Description
|
|
Genome
|
All the DNA chromosomes
|
|
22
|
Autosomal chromosomes
|
|
2
|
Sex chromosomes
|
|
Females
|
XX
|
|
Males
|
XY
|
|
genes
|
Coding regions
|
|
~30,000 genes
|
~5% in genomic DNA
|
|
Locus
|
Gene location
|
|
Homozygous
|
DNA sequences in two alleles at a genetic locus on homologous chromosomes are similar.
|
|
heterozygous
|
DNA sequences in two alleles at a genetic locus on homologous chromosomes are different.
|
|
Genotype
|
Genotype refers to alleles that are present at a genetic locus.
|
|
DNA profile
|
DNA profile refers to the combination of genotypes obtained for multiple loci.
|
|
DNA profiling
|
DNA profiling refers to the process of determining the genotype present at specific locations along the DNA molecule.
|
Reference
ATCC cell lines: https://www.atcc.org/en/Products/Cells_and_Microorganisms/Cell_Lines.aspx
Butler, J.M.; Forensic DNA Typing. 2nd Edition.2005 Elsevier Academic Press.
BSI blog
https://blog-biosyn.com/2013/04/15/why-perform-a-cell-identification-check/
https://blog-biosyn.com/2013/02/18/cell-line-authentication-and-identification/
https://blog-biosyn.com/2013/06/27/why-should-i-test-my-cho-cell-culture-for-virus-infections/
Cabrera CM, Cobo F, Nieto A, Cortés JL, Montes RM, Catalina P, Concha A.; Identity tests: determination of cell line cross-contamination. Cytotechnology. 2006 Jun;51(2):45-50. doi: 10.1007/s10616-006-9013-8. Epub 2006 Aug 3.
Chatterjee R.; Cell biology. Cases of mistaken identity. Science. 2007 Feb 16;315(5814):928-31.
https://www.ncbi.nlm.nih.gov/pubmed/17303729
DNA Analyst Training – Laboratory Training Manual Protocol 5.02 PCR: Amplification and Electrophoresis of STRs. Presideint’s DNA Initiative.
https://static.training.nij.gov/lab-manual/Linked%20Documents/Protocols/pdi_lab_pro_5.02.pdf.
Drexler HG, Dirks WG, MacLeod RA.; False human hematopoietic cell lines: cross-contaminations and misinterpretations. Leukemia. 1999 Oct;13(10):1601-7.
Hans G. Drexler, Roderick A. F. MacLeod, Willy G. Dirks; Cross-contamination: HS-Sultan is not a myeloma but a Burkitt lymphoma cell line. Blood 2001 98:3495-3496; doi:10.1182/blood.V98.12.3495
Hashiyada, M.; DNA Biometrics. www.intechopen.com http://cdn.intechopen.com/pdfs/16506/InTech-Dna_biometrics.pdf
Masaki Hashiyada (2011). DNA biometrics, Biometrics, Dr. Jucheng Yang (Ed.), ISBN: 978-953-307-618-8, InTech, Available from: http://www.intechopen.com/books/biometrics/dna-biometrics
MacLeod RA, Dirks WG, Matsuo Y, Kaufmann M, Milch H, Drexler HG.; Widespread intraspecies cross-contamination of human tumor cell lines arising at source. Int J Cancer. 1999 Nov 12;83(4):555-63.
Roderick A.F. MacLeod, Wilhelm G. Dirks and Hans G. Drexler; One falsehood leads easily to another. Int. J. Cancer: 122, 2165-2168 (2008).
User Guide - AmpFιSTR® Identifiler® PCR Amplification Kit ABI – life technologies (PN: 4322288).
-.-