Zip Nucleic Acids (ZNA®) Oligos, Oligospermine-oligonucleotide conjugates, a potent oligo modification to be used in in molecular biology, diagnostic and therapeutic applications. The central point of these applications is the ability of the nucleobases to form Watson-Crick base-pairing, whereby complementary strands form duplexes with each other. However, the strength of such interactions is tempered by the mutual electrostatic repulsion of the negatively charged phosphate backbones running through each strand. This polyanionic nature of
oligonucleotides also hinders their ability to enter cells and access therapeutically interesting targets that are in the cell cytosol or nucleus.
Numerous research on nucleobase analogs focus to address the issue of of electrostatic repulsion to improve both the binding of an oligonucleotide to a target sequence and its cell delivery. Several approaches aimed at reducing the anionic charge can be found in the literature ranging from the replacement of the phosphate backbone with one that is amide based, such as in Peptide Nucleic Acids (PNA),1 or the introduction of ammonium or guanidinium residues throughout the strand,2,3 to the conjugation of polycations that preserve the hybridization specificity and nucleic acid-processing enzyme activity of the parent oligonucleotide.4 In the case of the latter, a convenient way of introducing a polycation polymer is by the use of the spermine nucleobase (Figure 1) during oligo synthesis.5,6 The resulting oligonucleotide-oligospermine conjugates (illustrated above and in Figure 2) are known as “Zip Nucleic Acids” or “ZNA®”,7 a term that reflects the presumed mode of action of the conjugates that are believed to use the oligospermine to seek out and move along (scan) oligonucleotide strands until the probe complementary sequence is located. The oligospermine then performs the function of stabilizing the formed duplex by reducing electrostatic repulsion, thereby leading to significantly increased binding affinities. Despite this increase, ZNA® oligonucleotides still retain the ability to discriminate mismatches, doing so to the same extent as unmodified DNA.8 Moreover, the oligospermine can be located at the 3’ or 5’ terminus depending on the application, producing the same beneficial effects to the binding affinity.
Figure 1: Structure of Spermine Nucleobase
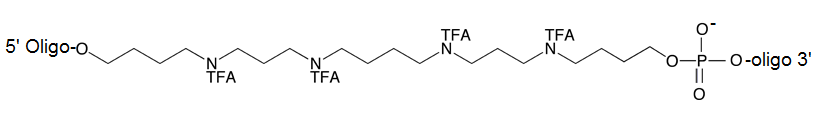
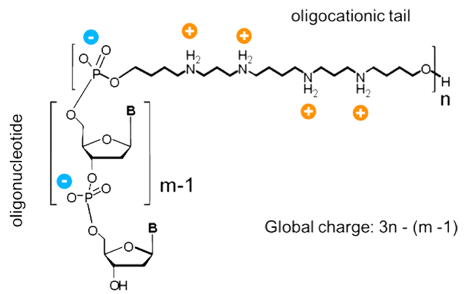
Figure 2: Oligonucleotide Oligospermine Conjugate

Depiction of ZNA® molecule whereby the oligospermine tail is attached to the 5’ of a DNA oligomer. The net charge is determined by the formula 3n-(m-1), where m is the oligonucleotide length and n the number of spermine units.
Figure 3: Comparative PCR Performance of ZNA ZNA® and DNA primers on an AT-rich Sequence
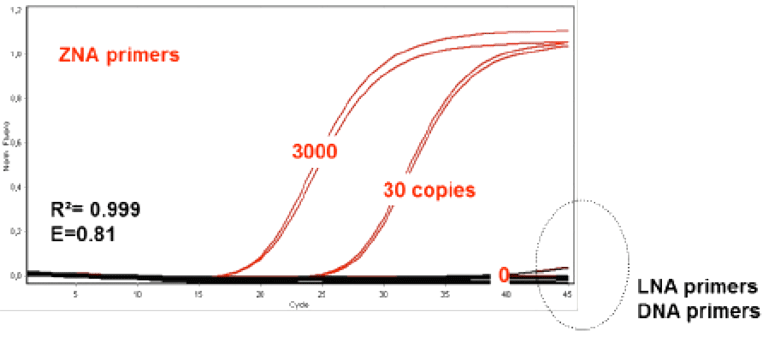
Figure 3: ZNA® primers (Red) used for AT-rich regions in comparison to the corresponding LNA and DNA primers (Black) at 100nM primer concentration and varying copies of initial template following a standard 2 step protocol (95°C, 10s followed by 60°C, 30s)
ZNA® oligos also have notable advantages when used as primers for reverse transcription (RT).9 In this respect, it was shown that ZNA® primers improve the yield of RNA to cDNA conversion under standard conditions that led to earlier detection in RT-qPCR quantifications compared to DNA. As a result, this leads to more accurate quantification of low-abundant transcripts. Moreover, the primers in such applications were found to be more robust to variations in buffer environment compared to unmodified DNA primers.
The properties of ZNA® molecules make them ideal for use as dual-labelled hydrolysis probes for qPCR. 3’-attached oligospermine-oligonucleotide conjugates can be made shorter than their non-modified counterparts leading to improved single-nucleotide polymorphism discrimination (Figure 4).8 In addition, longer ZNA® of a length similar to that used for non-modified probes, exhibited reduced background fluorescence through increased quenching, thus generating improved signal-to-noise ratios. Such a finding opens the way for ZNA® to be used as non-hydrolysis probes. Due to the predictable increases in Tm afforded to the molecules by the attached spermine units, it is easier to design ZNA® probes compared to alternative oligonucleotide modifications such as minor groove binders (MGB) or LNA probes. Moreover, it was found that ZNA® tolerated a wider range of conditions than their DNA and LNA counterparts. In a recent report by Trevisan and coworkers,10 ZNA® probes were successfully used for the in-situ detection of microRNAs in plants. The probes were shown to be highly sensitive and selective when compared to LNA probes and indicate a potential for exploitation of ZNA® for high-throughput miRNA profiling applications.
Figure 4: Improved single-nucleotide polymorphism (SNP) discrimination with short ZNA® probes
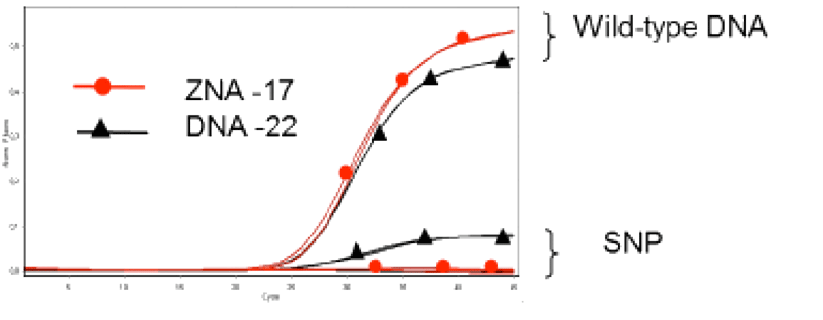
A 17mer DNA conjugated with 4 spermines (Red) shows better mismatch discrimination compared to an unmodified DNA 22mer (Black).
ZNA® Oligos as antisense and antigene agents
As the efficiency of cell-uptake of oligonucleotides is improved by their conjugation to cationically charged moieties,11 it follows that polyspermine modification also renders ZNA® more amenable to cell delivery. This point has been exploited, for example, by the group of David Corey who assessed the use of ZNA® as antisense and antigene agents.12
In-vitro assays revealed that oligospermine modification of control DNA and LNA/DNA mixmers had a significant beneficial impact on antisense activity when delivered with cationic lipid. For instance, a previously nonfunctional DNA molecule attained antisense activity through the addition of 6 spermine units. More interestingly, free delivery (without cationic lipid mediated transfection) yielded IC50 values in the mid-nanomolar range (Table 1).
Table 1: Antisense Activity of ZNA®oligonucleotides
|
|
IC50 (nM) with
cationic lipid
|
IC50 (nM) without
cationic lipid
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Antisense Activity of ZNA®oligonucleotides in the presence and absence of cationic lipid.12 Spermine (Z) additions to DNA (gctgctgctgctgctgctg) directed towards the CAG repeat of mutant huntingtin (HTT) mRNA triggered antisense activity with and without cationic lipid mediated transfection. The same sequence with LNA substitutions for Thymidine, LNA(T), also showed significant improvement in assisted and free delivery
|
A similar outcome was seen with ZNA® oligos used for the antigene application. Oligospermine-ribonucleotide conjugates have also been reported as a way to transport siRNA into cells. Nothisen and coworkers13 synthesized several 5’-oligospermine-sense strand conjugates and determined that an siRNA net cationic charge was necessary to effect efficient cell uptake and gene silencing in the submicromolar range. These encouraging results were also accompanied by an indirect confirmation of the siRNA mechanism, namely that switching the oligospermine to the 5’- of the antisense strand resulted in loss of activity.
Intellectual Property
"Spermine phosphoramidite" synthon is the subject matter of U.S. Patent Application No. 12/086.599, European Patent Application No. EP20060847298 and foreign equivalents for which Polyplus-transfection is the co-owner. Product is sold for research purposes only.
References:
- Nielsen, P.E. et al. Science 1991, 254 (5037), p.1497–1500.
- Michel, T. et al. Nucleic Acids Res. 2003, 31, p. 5282-5290.
- Barawkar, D.A.; Bruice, T.C. Proc. Natl. Acad. Sci. USA 1998, 95, p.11047-11052.
- Fraley, A.W. et al. J. Am. Chem. Soc. 2006, 128, p.10763-10771.
- Pons, B. et al. ChemBioChem 2006, 7, p1173-1176.
- Voirin, E. et al. Nature Protocols 2007, 2, p.1360-1367.
- Noir, R. et al J. Am. Chem. Soc. 2008, 130, p.13500-13505.
- Paris, C. et al. Nucleic Acids Res. 2010, 38, e95.
- Moreau, V. et al. Nucleic Acids Res. 2009, 37, e130.
- Trevisan, S. et al. Plant, Cell & Environment 2012, 35, p. 1137-1155.
- Juliano, R. et al Nucleic Acids Res. 2008, 36, p.4158–4171.
- Gagnon, K.T. et al. J. Am. Chem. Soc. 2011, 133, p.8404-8407.
- Nothisen, M. et al. J. Am. Chem. Soc. 2009, 131, p.17730-17731.