Abstract
Mutations on epidermal growth factor receptor (EGFR) cause a variety of cancers including breast and lung cancers. The single mutation T790M on tyrosine kinase domain of EGFR signifies the response to the popular cancer drug gefitinib, which leads to the development of resistance to gefitinib. Detecting the mutation thus guide effective therapeutical options for patients who are in need of cancer drug treatments. We sought to develop a rapid, reliable detection method for the T790M mutation using bridged nucleic acids (BNA), which has been known to enhance the hybridization affinity of oligonucleotides that contain BNA bases. Oligonucleotides containing BNA bases designed to block PCR reaction against wild-type genes, called BNA-clamp, were used to discriminate the presence of mutant genes mixed with a large number of wild-type genes. Real-time PCR in conjugation with BNA-clamping allows us to view the different levels of PCR amplifications in the degree of mixture of wild-type and mutant genes. In an effort to explore the possibility, 13-mer long clamps were prepared with various numbers of BNA bases. The clamps containing 9 BNA bases appear to be most effective in blocking the PCR reactions at an optimized concentration to distinguish the mutant from the wild-type genes. In addition to PCR results, the deferencein the Tm values for the 7 BNA bases in the 13-mer was the largest among differently designed clamps, which is consistent with the results obtained by real-time PCR. We also examined the degree of sensitivity using the clamp containing 9 BNA bases, revealing that the clamp has the ability to determine the level of mutation as low as 1% mixture of the mutant and wild-type gene. The effectiveness of blocking the PCR reaction with only 13-mers containing BNA bases allowed us to detect a single mutation, and thus, this BNA-clamping real time PCR technology may offer a promising, new avenue to detect clinically important mutations in the future.
Introduction
The receptor tyrosine kinase epidermal growth factor receptor (EGFR) plays a vital role in signal pathways including cell proliferation, migration, metastasis, evasion of apoptosis, and angiogenesis. Mutations on the EGFR, however, lead to overexpression of the receptor causing a number of cancers such as lung cancer and glioblastormamultiforme. To treat the overexpression of EGFR, gefitnib has been used as a drug. The use of gefitnib has unfortunately resulted in the emergence of resistance to gefinib by the mutation T790M located at exon-20 of EGFR (Figure 1). It suffices to say that one-half of the patients with the mutation eventually anguish with the cancer recurrence. Thus, if it would be possible to detect the T790M mutation, it would provide a better choice for cancer drugs.
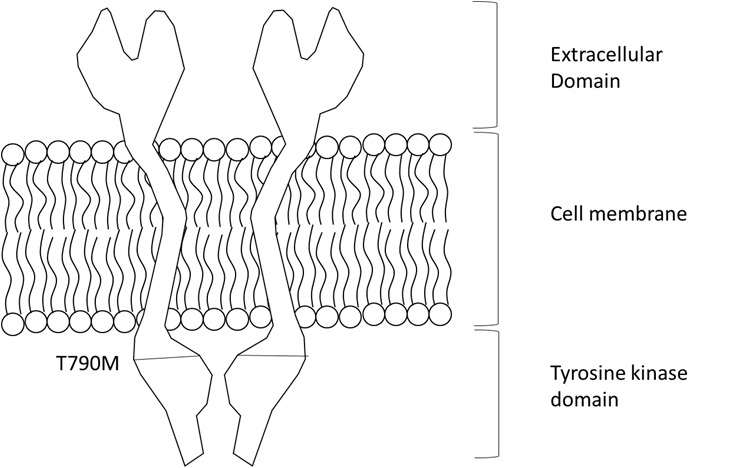
Figure 1. A cartoon version of EGFR and the T790M location of EGFR. The T790M mutation is located at the tyrosine kinase domain, inducing activation of signaling.
To detect the mutations, highly sensitive method is required. The most promising and developing method is polymerase chain reaction (PCR) clamp technology, which has been employed as a rapid, sensitive method for the detection of gene mutations.
We have developed an easy, cost-effective, and rapid method to detect the single point mutation T790M that was synthetically generated. We have explored the possibility of a BNANC-clamp-based real-time PCR using SYBR green dye, where BNANC stands for the 3rd generation bridged nucleic acid (Figure 2). A series of BNA-clamps have been tested to understand their ability to discriminate the mutant in the large quantity of wild-type genes. The Tm values of BNA-clamps was also determined.
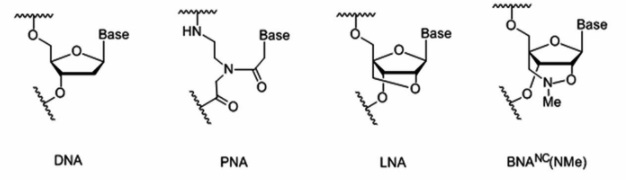
Figure 2. The structures of DNA and synthetic nucleic acids. The first one is the structure of DNA. The second one is the structure of PNA, peptide nucleic acid. The third one is the structure of LNA, locked nucleic acid. This is also known as the 1st generation BNA, 2’4’-BNA. The last one is the structure of BNANC, which is also known as the 3rd generation BNA.
Methods
BNANC-clamp real-time PCR
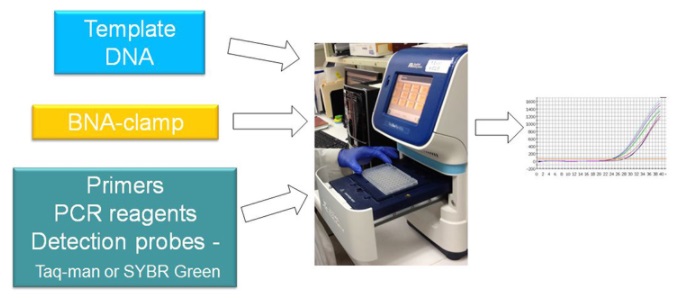
Results
Table 1. The BNANC-clamping oligonucleotides and Tm values of the wild-type exon-20 gene and the mutant T790M of exon-20.
| Name |
Length |
# of BNA |
Tm |
| WT |
T790M |
DTm |
| BNANC-clamp-5 |
13 |
5 |
70.9 |
65.1 |
5.8 |
| BNANC-clamp-7 |
13 |
7 |
75.1 |
68.6 |
6.5 |
| BNANC-clamp-9 |
13 |
9 |
82.1 |
72.1 |
10.0 |
| BNANC-clamp-11 |
13 |
11 |
86.3 |
80.2 |
6.1 |
| BNANC-clamp-13 |
13 |
13 |
N.D.* |
N.D. |
- |
*N.D. represents that the Tm value cannot be determined.
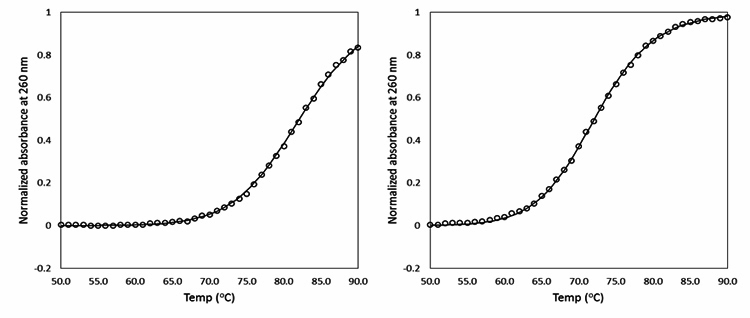
Figure 3. Tm values of BNANC-clamp-9 against WT and T790M mutant genes. The left panel shows the data of oligonucleotide denaturation between the WT DNA and the BNANC-clamp9. The right panel shows the data of denaturation between T790M mutant and the BNANC-clamp9.
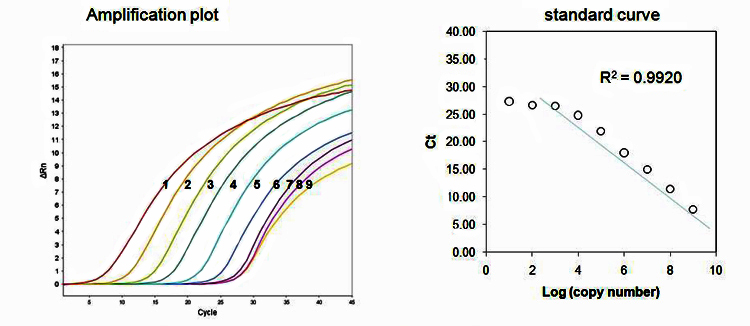
1: 109 copies/PCR; 2: 108 copies/PCR; 3: 107 copies/PCR; 4: 106 copies/PCR; 5: 105 copies/PCR; 6: 104 copies/PCR; 7: 103 copies/PCR; 8: 102 copies/PCR; 9: 101 copies/PCR
Figure 4. Detection limit analysis. The left panel shows the real-time PCR amplification results depending on the copy number of gene. The gene used is the plasmid DNA containing WT exon-20. The right panel shows the plot of the copy numbers vs. Ct values.
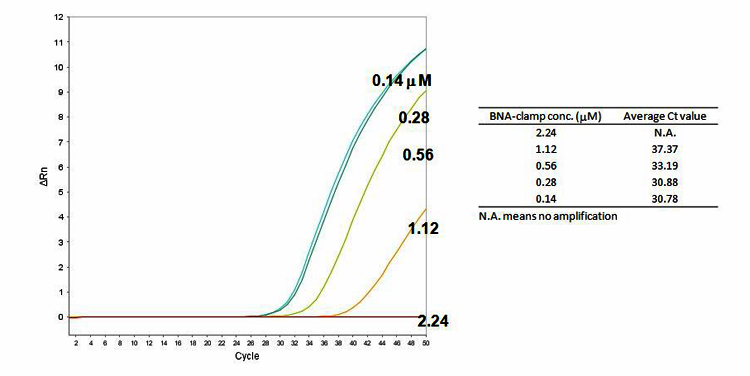
Figure 5. Effectiveness of BNANC clamping for plasmid DNA. 104 copy number of WT plasmid DNA was used
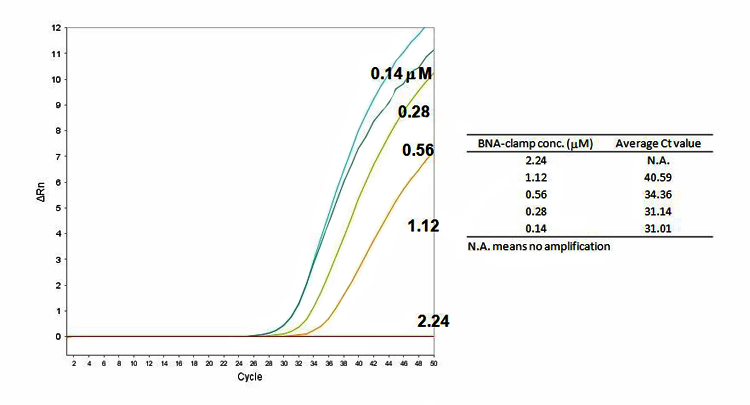
Figure 6. Effectiveness of BNANC clamping for genomic DNA. 104 copy number of WT genomic DNA was used.
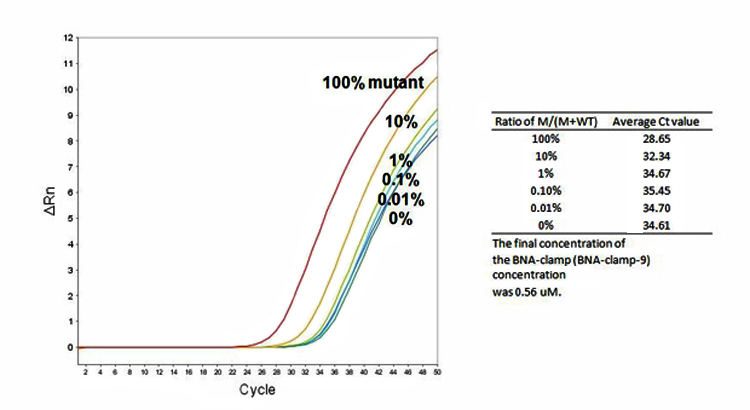
Figure 7. Sensitivity test results using 104 copy number of T790M plasmid DNA and various concentration of WT genomic DNA
Conclusions
-
In a series of BNANC clamps listed in Table 1, we found that BNANC-clamp9 containing nine BNANC bases shows the largest difference in the Tm values between WT and the mutant T790M.
-
The detection limit shows the possibility of amplification down to 103 copy number of gene.
-
The BNANC-clamp9 is able to suppress the gene amplifications by the concentration of BNA clamp.
-
In comparison between plasmid DNA and genomic DNA, no significant difference was observed in real-time PCR amplification.
-
BNANC-clamp9 is able to determine the level of mutation as low as 1% mixture of the mutant and wild-type gene.
Bibliography
- Rahman, S.M.; Seki, S.; Utsuki, K.; Obika, S.; Miyashita, K.; Imanishi, T., “2',4'-BNA(NC): a novel bridged nucleic acid analogue with excellent hybridizing and nuclease resistance profiles”, Nucleosides Nucleotides Nucleic Acids., 2007, 26, 1625-1628.
- Rahman, S. M.; Seki, S.; Obika, S.; Yoshikawa, H.; Miyashita, K.; Imanishi, T., “Design, synthesis, and properties of 2',4'-BNA(NC): a bridged nucleic acid analogue”, J. Am. Chem. Soc., 2008, 130, 4886-4896.