Chemical solid phase and solution phase synthesis methods allow the synthesis of circular oligonucleotides, both for the production of circular DNA and RNA molecules. However, the use of enzymatic approaches may of advantage in some cases. Small circular single-stranded oligonucleotides (<28 base pairs {bp}) can be chemically synthesized using solution and solid-phase methods from partially protected linear precursor molecules. Longer circular oligos (>28 bp) are relatively easy to synthesize by both methods, chemical and enzymatic. The chemical circularization of linear oligonucleotides can be achieved using cyanogens bromide (BrCN) or the carbodiimide cross-linker 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) together with a template functioning as a bridge. However, small circular oligonucleotides, for example containing 28 to 50 bp, are readily synthesized using a single precursor segment. However, large-sized circles, containing more than 50 bp, can be prepared utilizing two or three smaller precursor oligonucleotide segments. Unfortunately, the preparation of large circular oligonucleotides from multiple precursor segments often results in low yields. In addition to chemical circularization methods, enzymatic ligation offers an alternative approach for the preparation of circular oligonucleotides. For this approach, T4 ligases are routinely utilized for the cyclization reactions.
For the enzymatic synthesis of circular DNA and RNA oligonucleotides, T4 ligases are commonly used. These enzymes can circularize the reactive 3’-hydroxyl (OH) and 5’-phosphate (PO4) groups of linear ssDNA using a complementary short template. The enzyme DNA ligase catalyzes the covalent bond formation between the 3’OH and the 5’PO4 on DNA phosphodiester bonds. The reaction requires two ends of double-stranded DNA and ATP. Bacteriophage T4 ligase is the enzyme of choice most often used for because it can ligate blunt-ended DNA as well as DNA with compatible cohesive ends. However, the optimal temperature may vary for different reaction conditions. T4 RNA ligase I can circularize linear ssRNA in the absence of a template.
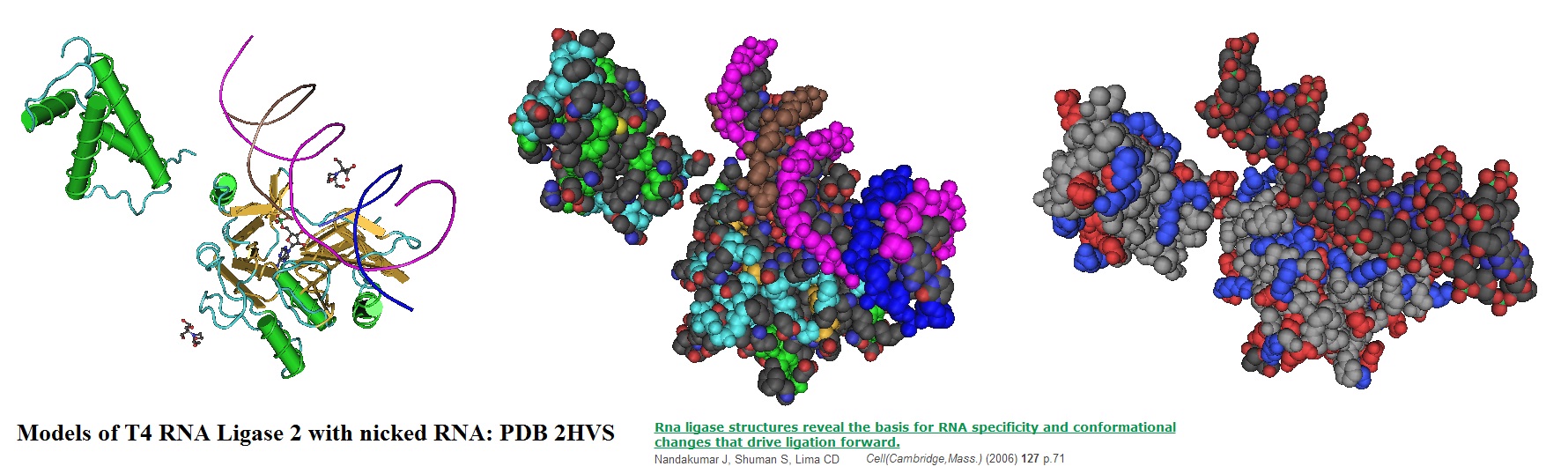
Figure 1: Models for the T4 RNA ligase 2 with nicked RNA. A synthetic construct of Enterobacteria phage T4 was used to generate crystal structures. The resulting structural models illustrate the stereochemistry of the nucleotidyl transfer. In addition, the remodeling of active-site contacts and conformational changes that propel the ligation reaction forward are revealed (Nandakumar et al., 2006).
T4 RNA ligase 2 (Rnl2) and kinetoplastid RNA editing ligases belong to a family of RNA repair enzymes. RNA ligases seal 3'-OH/5'-PO(4) nicks in duplex RNAs via ligase adenylylation, followed by a transfer of an adenosine monophosphate (AMP) to the nick 5'PO(4) group. The attack by the nick 3'-OH on the 5'-adenylylated strand forms a phosphodiester bond.
Resulting products can be separated and visualized using denaturing polyacrylamide gel electrophoresis (PAGE). The slower migration mobility of circular oligonuleotides in comparison to their linear counterparts circular products allows the observation of these products as stained bands migrating with higher apparent molecular weights (figure 2). In addition, success of the circularization reaction can be determined with the help of endonuclease cleavage.
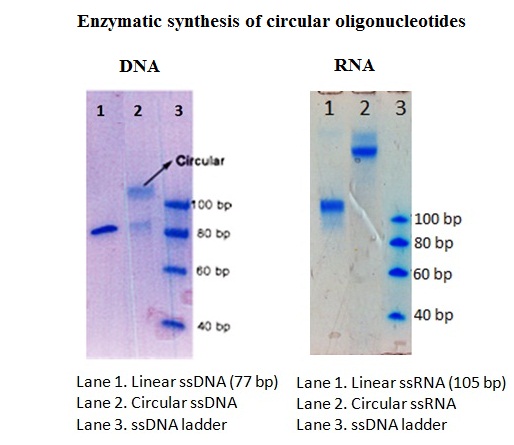
Figure 2: Quality control of enzymatic synthesis products of circular DNA and RNA using PAGE.
References
Beaudry D., Perreault J-P. An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Research, 1995, Vol. 23, 3064-3066. [PMC]
Diegelman A. M. and Kool E. T. Chemical and enzymatic methods for preparing circular single-stranded DNAs. Current Protocols in Nucleic Acid Chemistry, 2000, 5.2.1-5.2.27. [Current Protocols]
Dolinnaya NG, Blumenfeld M, Merenkova IN, Oretskaya TS, Krynetskaya NF, Ivanovskaya MG, Vasseur M, Shabarova ZA. Oligonucleotide circularization by template-directed chemical ligation. Nucleic Acids Res. 1993 Nov 25;21(23):5403-7. [PMC]
Nandakumar J, Shuman S, Lima CD; Rna ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell(Cambridge,Mass.) (2006) 127 p.71.
Pedroso E., Escaja N., Frieden M., and Gradas A. Solid-phase synthesis of circular oligonucleotides. Methods in Molecular Biology, Vol. 288, 2005: Oligonucleotide synthesis: Methods and Applications. Edited by: P. Herdewijn, Humana Press Inc., Totowa, NJ Page 101-125. [PubMed]
---...---