Modified synthetic oligonucleotides can be used to probe and study epigentic events. Bio-Synthesis provides custom synthesis of oligonucleotides containing methylated DNA bases for epigenetic studies. Epigenetics is the study of changes in heritage control of gene expression through an identical DNA sequence, yet maintains different terminal phenotypes. This nongenetic cellular memory, which records developmental and environmental cues as well as alternative cell states in unicellular organisms, is the basis of epigenetics. The growing interest to fully explain the heritability of complex traits, and to pinpoint the genetic effects in some complex diseases, along with the desire to understand why cells have “deprogramming” into pluripotent/totipotent states, has led epigenetic research into many regulatory systems involving switching a gene from its 'on' to the 'off' state or vise versa in areas of DNA methylation, histone modification, nucleosome location, and noncoding RNAs. Until 2009, the knowledge about DNA modifications was limited to the introduction of a methyl group at the C5 position of the cytosine base, which converts 2’-deoxyCytidine (dC) into 5-methyl-2’-deoxyCytidine (mdC).
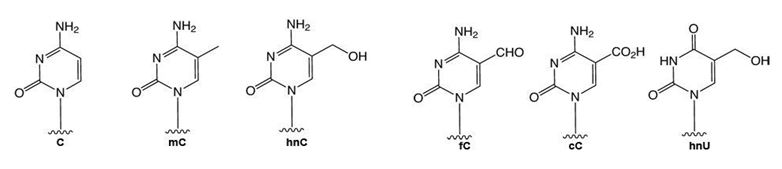
Figure 1: Structures of modified cytidines.
C = cytidine, mC = 5-methylcytidine, hnC = 5-hydroxymethylcytidine, fC = 5-formylcytidine, cC = 5-carboxylcytidine, and hmU = 5-hydroxymethyluridine.
This methylation occurs predominantly in CpG islands (areas with high occurrence of the CG motif) of promoters. mdC is introduced by specialized DNA methyltransferase (DNMT) enzymes.
A study in 2009 reported (1,2) the discovery of 5-hydroxymethyl-2’-deoxyCytidine (hmdC), a novel dC modification in Purkinje neurons and embryonic stem cells. Later, a third report found this modification to be strongly enriched in brain tissues associated with higher cognitive functions (3). This new dC modification is generated by the action of α-ketoglutarate dependent TET enzymes (ten eleven translocation), which oxidizes mdC to hmdC. This finding stimulated discussion about active demethylation pathways that could occur, for example, via base excision repair (BER), with the help of specialized DNA glycosylases. Alternatively, one could envision a process in which the hydroxymethyl group of hmdC is further oxidized to a formyl or carboxyl functionality followed by elimination of either formic acid or carbon dioxide (4,5) .
A number of recent publications provides data that support both pathways. It was discovered that hmdC could be deaminated by activation-induced deaminase (AID) enzymes to provide 5-hydroxymethyl-2’-deoxyUridine (hmdU). This compound was shown to be excised by the SMUG-1 DNA glycosylase (6). After initial failure to detect any further oxidized hmdC derivatives in somatic tissues (4), newly developed mass spectrometric technologies, in combination with the available reference compounds, finally enabled researchers to gather strong support for the putative oxidative demethylation pathway. These methods and standards enabled the discovery of 5-formyl-2’-deoxyCytidine (fdC) in differentiating embryonic stem cells (7). More recently, a similar technology also led to the discovery of 5-carboxyl-2’-deoxyCytidine (cdC), but the amount of fdC and cdC measured differs largely in all three reports (8)..
Research is currently ongoing to unravel the true levels and fate of these further oxidized dC bases in somatic tissues and in different stem cells. Even along the oxidative pathway, base excision processes have been proposed to play a major role with two reports showing that thymidine DNA glycosylase (TDG) accepts both fdC and cdC as substrates (8,9). A possible oxidative demethylation pathway would clearly rely on the existence of a dedicated decarboxylase that is able to convert cdC back into dC. Such an intriguing decarboxylation would enable nature to remove the 5-methyl group in mdC without introducing DNA strand-breaks that accompany any BER based base removal.
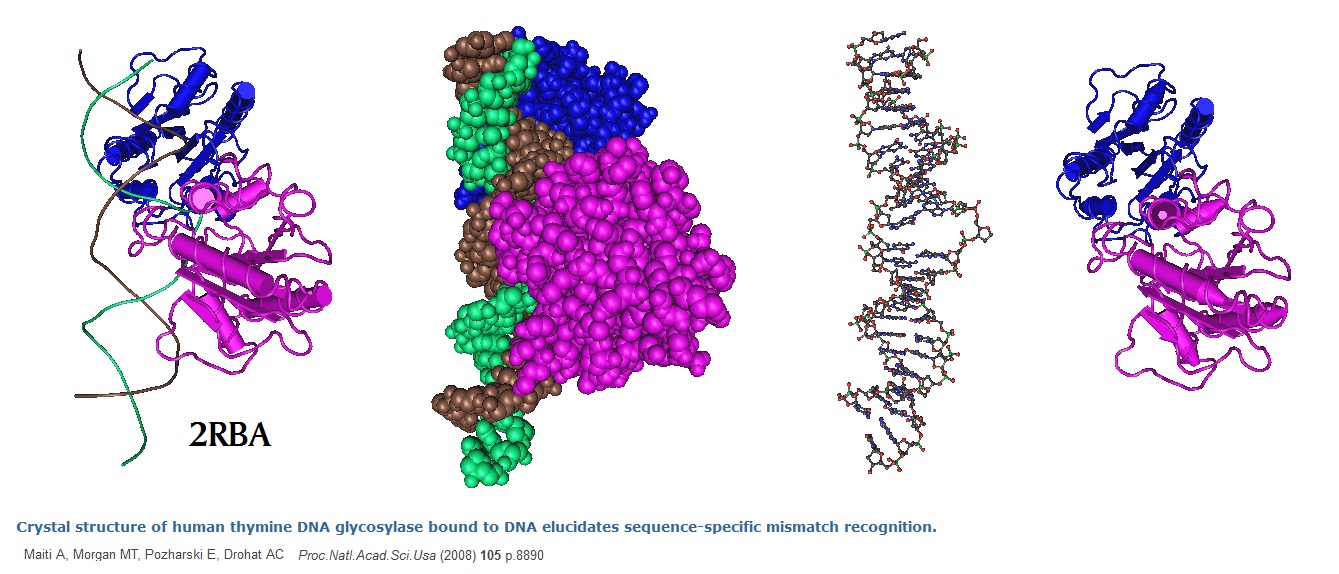
Figure 1: Structure of human thymine DNA glycosylase (TDG) bound to DNA. The epigentic modification m5CpG found on methylated CpG dinucleotides is important for transcriptional regulation and genomic stability in vertebrate cells. m5C deamination yields G-T mispairs. These mispairs are implicated in gentic diseases such as cancer as well as in aging. Human thymine DNA glycosyalse (hTDG) can remove thymine (T) from G-T mispairs. This enzymatic reaction produces abasic (AP) sites. Special repair proteins restore the G-C pair in a follow-on reaction.However, hTDG is inactive against A-T pairs but is most effective for G-T mispairs and other damage located in a CpG region. The crystal structure reported by Maiti et al. in 2008 revealed molecular interactions that promote the stringent specificity needed for guanine versus adenine as the pairing partner of the target base. The observed interactions most likely confer CpG sequence specificity (See Maiti et al. 2009 for more info).
Bio-Synthesis is supporting epigenetic resarch by providing synthetic oligonucletoides containing all the new cytosine analogues: hmdC, fdC and cdC. These modified bases can be incorporated during synthesis at any position using conventional solid-phase oligonucleotide synthesis chemistry by replacing the standard cytosine DNA base with methylated DNA cytosine base.
- 5-hydroxymethyl-dC [hmdC]
- 5-hydroxymethyl-dC II [hmdCII]
- 5-carboxy-dC [cdC]
- 5-Formyl-dC [fdC]
The first generation hmdC phosphoramidite was fairly very well accepted but requires fairly harsh synthesis conditions. Therefore, a second generation building block (5-Hydroxymethyl-dC II) developed by Carell and co-workers that is compatible with UltraMild deprotection has been introduced (6). 5-Formyl-dC and 5-carboxy-dC may find uses in research into DNA damage and repair processes.
References:
-
S. Kriaucionis, and N. Heintz, Science, 2009, 324, 929-30.
-
M. Tahiliani, et al., Science, 2009, 324, 930-935.
-
M. Münzel, et al., Angewandte Chemie-International Edition, 2010, 49, 5375-5377.
-
D. Globisch, et al., PLoS One, 2010, 5, e15367.
-
S.C. Wu, and Y. Zhang, Nat Rev Mol Cell Biol, 2010, 11, 607-20.
-
M. Münzel, D. Globisch, C. Trindler, and T. Carell, Org Lett, 2010, 12, 5671-3.
-
M. Münzel, et al., Improved Synthesis and Evaluation of Oligonucleotides Containing 5-Hydroxymethylcytosine, 5-Formylcytosine and 5-Carboxylcytosine. In Chemistry - A European Journal, 2011, in press.
-
Atanu Maiti and Alexander C. Drohat; Thymine DNA Glycosylase Can Rapidly Excise 5-Formylcytosine and 5-Carboxylcytosine. THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 286, NO. 41, pp. 35334–35338, October 14, 2011.
-
Maiti A, Morgan MT, Pozharski E, Drohat AC; Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proc.Natl.Acad.Sci.Usa (2008) 105 p.8890.