PEGylation of Therapeutic Proteins
Therapeutic proteins such as monoclonal antibodies and cytokines, after delivery to the body, are eliminated by proteolysis and clearance in the kidney or liver, a process that limits their usefulness as therapeutic agents. In addition, many of these proteins can also trigger an antibody immune response that would block their beneficial effects. Hence, significant efforts have been dedicated developing methods that while extending these proteins’ circulating life and decreasing their immunogenicity do not interfere with their specific biological functions. Between the different methods explored, pegylation or the covalent addition of polyethylene glycol (PEG) chains to a protein, has been the most successful. Pegylation is used in several types of therapeutic proteins. Apparently it results in the increase of the hydrodynamic volume of a protein. However, addition of PEG can also have beneficial effects on nucleic acids by increasing their stability against nucleases.
PEGylation, or pegylation, refers to the covalent attachment of polyethylene glycol polymer chains to molecules such as proteins, peptides, oligonucleotides or other biomolecules.
The first FDA-approved pegylated therapeutic protein appeared on the market in 1990, which was Adagen®, a PEGylated form of adenosine deaminase. Following that, seven successful pegylated therapeutic proteins have been approved by the FDA, including Oncaspar®(Pegaspargase), PEGIntron ® (PEGylated IFN-α2b), Pegasys® (PEGylated IFN-α2a), Neulasta® (pegfilgrastm), Somavert® (Pegvisomant), Mircera® (PEGylatedepoetin-β), and Cimzia®(Certolizumabpegol). In addition to these already approved PEGylated biopharmaceuticals, many new products are currently under investigation and in different stages of clinical trials. Still, when considering pegylation as a strategy to extend the circulating life of a protein, several factors need to be considered, which will be discussed here.
-
Location of the PEG chains:
As a rule, PEG chains should be covalently linked at a site distant from the binding site of the protein, i.e. that region of the protein that is involved in binding to a cell receptor or an antigen; this way, the blocking of specific groups needed for binding and/or the steric hindrance of the binding process caused by the PEG are avoided. That means that at a minimum we should have amino acid sequence information about the protein as well as its binding site, post-translational modifications, such as glycosylation may be also helpful as they provide additional potential sites for the covalent binding of the PEG chains. Information about the tertiary structure of the protein would help in fine tuning the choice for a pegylation site. Indeed, a new approach is to modify the gene coding for a protein to add a new reactive group, like a cysteine or lysine, which is distal from the binding site, to allow the covalent binding of the PEG chain(s).
One strategy commonly used is to add the PEG chain at the N-terminal group of a protein, as usually this amino acid residue is not part of the binding site and it is located far from that site. Specificity for the terminal amino group is based on the differences in the pKa values for the lysine α and ε amino groups, and that the N-terminal group is always α. Another strategy used with glycosylated proteins is to oxidize the sugar moiety with periodate and to link the PEG to the aldehyde groups; because glycosylation sites are usually distant from the binding site, this approach minimizes any potential steric hindrance due to the added PEG.
-
Size and number of PEG chains:
In general, the larger the PEG chain the lower number of chains needed per mole of protein; yet, the length of the chain would be limited by its effects on stability and interference with binding. An advantage of longer PEGs is that usually a single chain will do, while smaller PEGs would require more chains added, which may interfere with the binding of the proteins. The size of the PEG is usually found by experimentation, in the case of monoclonal antibodies and their Fab fragments, PEGs of 40 kDaltons have been used successfully. However, while increase of the PEG size may result in a longer life, the biological activity of the protein may decrease as a function of the PEG size. Thus, the reason for the need to asses experimentally each therapeutic protein performance after pegylation. A new development in pegylation of therapeutic proteins is the use of branched lower molecular weight PEGs; these PEGs having usually two or three chains linked to a single linker carrying also a reactive group for binding to a protein, apparently has lower side-effects while maintaining a good stability. Also a higher circulating life of a pegylated protein may result in the accumulation of breakdown products that can damage the kidney; hence, as indicated above, pegylation effects need to be evaluated case by case.
-
Chemistry of the linking process:
A number of PEGs are available that have a single terminal reactive group capable of forming a covalent bond with some of the functional groups from proteins. For instance, PEGs having an aldehyde group can be used to preferentially add a PEG chain at the protein N-terminal group. This goal can be achieved by adjusting the pH of the reaction to have most of α amino groups deprotonated and thus able to react with the aldehyde. Other reactive groups are carboxyl, amine, maleimide, succimidyl and triazinyl, which are able to react with many of the functional groups available in proteins. To prevent damage to the protein, the reactions are carried under aqueous conditions, avoiding extreme pH conditions and using a concentration of activated PEG closer to the one needed to deliver the desired degree of pegylation. Once the pegylation of therapeutic proteins has taken place the different conjugates showing different degrees of substitution can be separated by ion exchange chromatography using a salt gradient to remove the excess of activated PEG and resolve the proteins with different degrees of pegylation. The pegylated therapeutic proteins are then concentrated, filter sterilized, and stored in PBS at 4oC. It may be possible to lyophilize the pegylated proteins in the presence of an excipient like mannitol or sorbitol.
-
Analysis for the Pegylation of therapeutic proteins:
Several methods can be used to assess the quality of the final product, such as SDS-PAGE, hydrophopic interaction chromatography (HIC), size exclusion chromatography (SEC), and matrix assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF-MS).
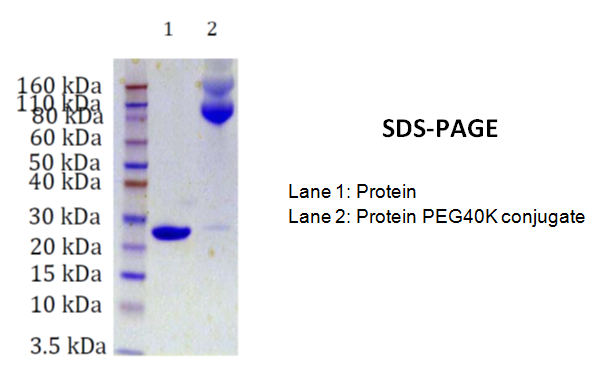
SDS-PAGE:
This method delivers information about the different conjugates prepared during the process. However, the molecular weight information is usually not related to the actual molecular weight.
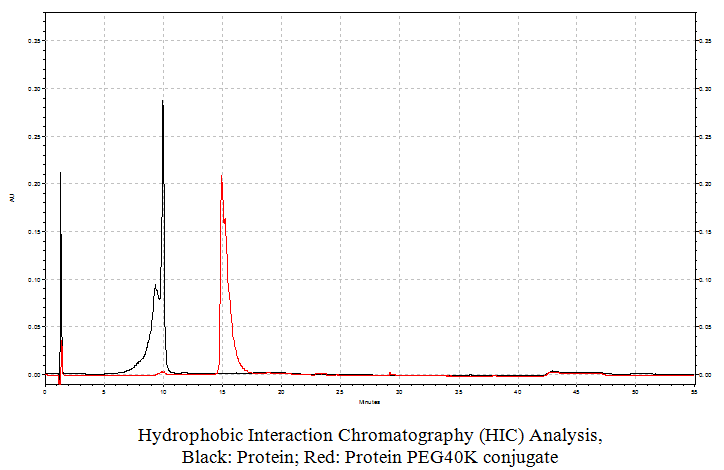
Hydrophobic Interaction Chromatography:
This method that allows resolving the different species according to their lipophilicity can be done using a butyl column, where the different conjugates are bound to the matrix using a high salt concentration and eluted in order of increasing lipophilicity by using a decreasing linear gradient of a salt, such as ammonium sulfate.
Size Exclusion Chromatography:
This method uses matrices like cross linked agarose gels, and the different conjugates are separated according to their hydrodynamic volume that is a result of the volume occupied by the protein plus the volume added by the PEG chain(s). This parameter is somewhat correlated with mol. wt. but it is not reliable to deliver the actual mol. wts.
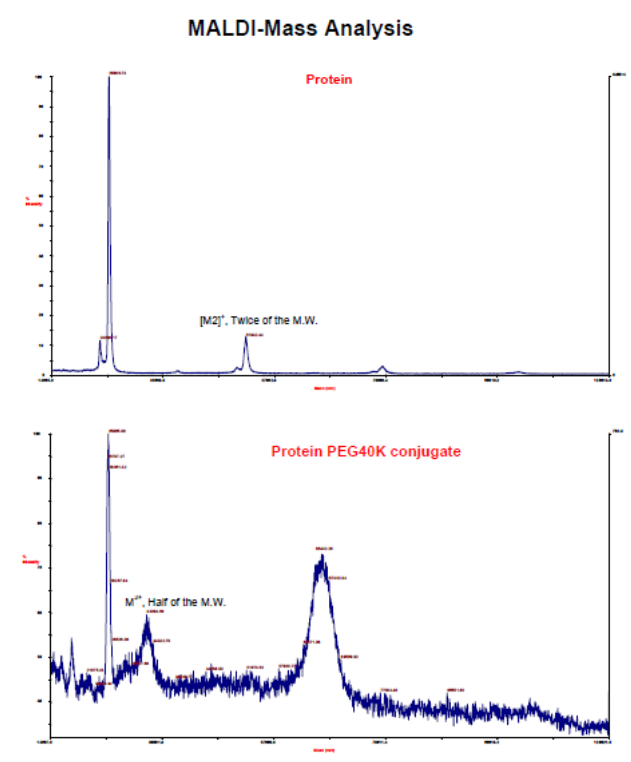
MALDI-Mass spectrometry:
Under optimal conditions, this method delivers the actual molecular weight of the conjugate and it is quite useful to establish the actual molecular weight of the pegylated proteins thereby indicating the presence of the correct pegylated protein conjugate.
Storage:
Pegylated proteins are best stored under sterile conditions at neutral pH and at 4oC. The buffer can be the common phosphate buffered saline solution at pH 7.2.
Reference
Ginn C, Khalili H, Lever R, Brocchini S.; PEGylation and its impact on the design of new protein-based medicines. Future Med Chem. 2014;6(16):1829-46. doi: 10.4155/fmc.14.125.
Jevsevar S, Kunstelj M, Porekar VG.; PEGylation of therapeutic proteins. Biotechnol J. 2010 Jan;5(1):113-28. doi: 10.1002/biot.200900218.
Veronese FM; Peptide and protein PEGylation: a review of problems and solutions. Biomaterials. 2001 Mar;22(5):405-17.
Yang T (2013) PEGylation – Successful Approach for Therapeutic Protein Conjugation. Mod Chem appl 1:e112. doi: 10.4172/2329-6798.1000e112