For experimental biomedical science, the ability of biotin to bind to streptavidin with high affinity has been exploited for both purification and detection purposes. This has also been the case with RNA molecules as labeling with biotin allows it to be immobilized on avidin-based supports for purification. As the biotinylated RNA could be detected by streptavidin coupled to chemiluminescent or fluorescent marker, it could also be used to detect the labeled RNA itself or its interacting target in various milieu.
The biotin labeling of RNA could be done site-specifically or randomly. To label RNA randomly, modified nucleotides could be incorporated during the in vitro transcription reaction. To label randomly in a single-step reaction, biotinylated UTP or biotinylated CTP can be incorporated during the co-transcriptional biotinylation step. The disadvantage with this method is that the labeling position(s) as well as the extent of labeling cannot be adequately controlled. Further, the biological function or property of the RNA may be altered or compromised through the biotin labeling.
This has led to the development of methods that enable site-specific labeling of RNA with biotin. To biotin label at a specific position located internally along RNA, a transcription-based method utilizing unnatural bases was developed (Moriyama et al., 2005). It relies on the ability of 2-amino-6-(2-thienyl)purine or 2-amino-6-(2-thiazolyl)purine to base pair with 2-oxo(1H)pyridine during transcription. The latter (conjugated to biotin) can be incorporated by T7 RNA polymerase during the transcription. A caveat to the method is that it requires one to prepare a template DNA containing unnatural bases beforehand.
To biotin label site-specifically at the 3' end of RNA, several methods are available. For chemical biotinylation, 3' terminal ribose of RNA can be oxidized to dialdehyde using periodate, which is susceptible to nucleophilic attack by amino group of a suitable biotin derivative (Willkomm and Hartmann 2005). Here, the disadvantage lies with the loss of 3' nucleotide through a competing reaction (beta-elimination) (Proudnikov and Mirzabekov 1996). Another method employs T4 RNA ligase to add a biotinylated pCp derivative to the 3' end of RNA (Kore et al., 2009). But the method is hampered by inefficient ligation reaction as well as the requirement for a high concentration of substrates. Other methods prescribed include the use of poly(A) polymerase to extend RNA at 3' end with N6-biotin-ATP; alternatively, phi29 DNA polymerase could be used to extend RNA after priming with an oligodeoxynucleotide (Moritz et al, 2013).
Various techniques have been devised to biotin label site-specifically at the 5' end of RNA. To selectively label at the 5' end (without simultaneously labeling the 3' end), RNA was incubated with aminopurine riboside triphosphate (to form a conjugate with RNA using T4 RNA ligase). The adenylated intermediate could then be labeled with biotin at the 5' end of RNA (Kinoshita et al., 1997). An alternate technique utilizes biotin-labeled nucleotide as the 'initiator nucleotide' for RNA transcription. To biotin label at the 5' end of RNA, a biotin-AMP conjugate (biotin-HDAAMP) can be used as transcription initiator under the T7 phi2.5 promoter (Huang et al., 2008) or biotinyl-guanosine 5'-monophosphate under the conventional T7 promoter (Collett et al 2005). Nevertheless, their main limitation is that neither method allows capping of the RNA.
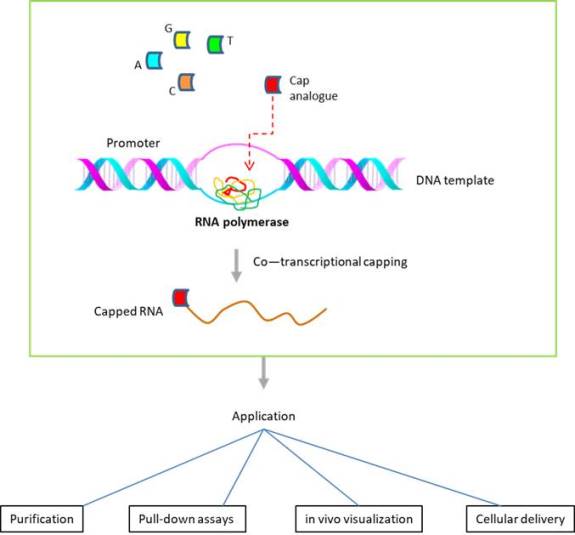
To address the above issue, additional approaches have been adopted. One method is based on 'co-translational capping', wherein a cap analogue is incorporated at the first nucleotide position (of the RNA transcript generated). The cap analogue could be further modified to incorporate in the correct orientation. To generate RNAs labeled with biotin via the 5' cap structure, the investigators at the University of Warsaw (Poland) developed biotin-labeled cap analogues modified within the triphosphate bridge, which could be incorporated for co-transcriptional capping (Bednarek et al., 2018).
Site-specific labeling of RNA molecules is a valuable tool with multiple applications, and Bio-Synthesis, Inc. provides site-specific biotin labeling of long RNA at 5’-end with nearly 100% efficiency.
References
Bednarek S, Madan V, et al. mRNAs biotinylated within the 5' cap and protected against decapping: new tools to capture RNA-protein complexes. Philos Trans R Soc Lond B Biol Sci. 373:20180167 (2018). PMID: 30397103
Collett,J.R., Cho,E.J., et al. Functional RNA microarrays for high-throughput screening of antiprotein aptamers. Anal. Biochem., 338, 113–123 (2005). PMID: 15707941
Huang F, He J, Zhang Y, Guo Y. Synthesis of biotin–AMP conjugate for 5′ biotin labeling of RNA through one-step in vitro transcription. Nat Protoc 3: 1848–1861 (2008). PMID: 18989262
Kinoshita, Y., Nishigaki, K., et al. Y. Fluorescence-, isotope- or biotin-labelinbeling of the 5-end of single-stranded DNA/RNA using T4 RNA ligase. 25:3747-8 (1997). PMID: 9278501
Kore AR, Charles I, et al. Synthesis and activity of modified cytidine 5′-monophosphate probes for T4 RNA ligase 1. Nucleosides Nucleotides Nucleic Acids 28: 292–302 (2009). PMID: 20183582
Moritz B, Wahle E. Simple methods for the 3' biotinylation of RNA. RNA. 20:421-7 (2014). PMID: 24448448
Moriyama K, Kimoto M, et al. Site-specific biotinylation of RNA molecules by transcription using unnatural base pairs. Nucleic Acids Res. 33:e129 (2005). PMID: 16113238
Proudnikov D, Mirzabekov A. Chemical methods of DNA and RNA fluorescent labeling. Nucleic Acids Res 24: 4535–4542 (1996). PMID: 8948646
Willkomm DK, Hartmann RK. 2005. 3′-Terminal attachment of fluorescent dyes and biotin. In Handbook of RNA biochemistry (ed. Hartmann RK, et al.), pp. 86–94. Wiley-VCH, Weinheim, Germany.