Template optimization for In Vitro Transcription
Bio-Synthesis provides enzymatic long RNA synthesis using transcription from DNA templates. In vitro transcribed RNA can be used as control template for Taqman SNP genotyping assays, drug metabolism genotyping assays or as positive control template for basic clinical research.
The development of an organism depends upon accurate and finely-tuned control of mRNA transcription. The round worm Caenorhabditis elegans (C. elegans) is a transparent nematode about 1 mm in length. This organism has been studied intensively in recent decades and has become a model organism for molecular biologists. Already various biological processes that involve direct regulation or specific functions of general transcription machinery components such as “Mediator”, or transcription co-factors that function in multiple discrete regulator systems, such as chromatin modification proteins, have been identified. For example, the multiprotein complex termed Mediator functions as a transcriptional coactivator in all eukaryotes. This protein complex was discovered by Roger D. Kornberg who was awarded the 2006 Nobel Prize in Chemistry for “his studies of the molecular basis of eukaryotic transcription". This multiprotein complex sometimes is also referred to as the Vitamin D Receptor Interacting Protein (DRIP) coactivator complex or the Thyroid Hormone Receptor-associated Proteins (TRAP). However, all genes have to be expressed in order to function in a cell. The first step in expression is transcription of the gene into a complementary RNA strand. For genes that code for transfer RNA (tRNA) and ribosomal RNA (rRNA) moleculs the transcript itself is the functionally important molecule. For other genes however, the transcript is translated into a protein.
The Central Dogma in Molecular Biology
The Central Dogma in molecular biology for prokaryotic in comparison to eukaryotic cells is outlined as follows:
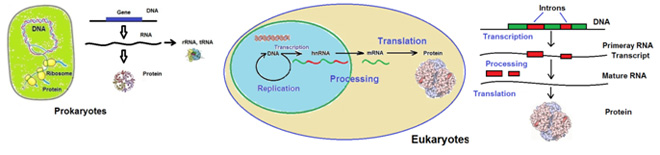
In prokaryotic cells, which have no nuclear membrane, DNA replication and transcription and RNA translation occurs in one compartment. Furthermore, the three processes can occur simultaneously.
In eukaryotic cells, which have a nuclear membrane, DNA replication and transcription occurs in the nucleus, and proteins are synthesized in the cytoplasm. RNA molecules therefore have to travel across the nuclear membrane before translation can happen. In these cells transcription and translation are physically separated. In addition, the primary transcript, heterogeneous nuclear RNA (hnRNA), is post-transcriptional processed to generate a messenger RNA (mRNA) molecule that can migrate through the nuclear membrane. The following figure illustrates this.
Advancements made in recent years in manual and automated oligonucleotide synthesis technologies allow now to routinely synthesize DNA, RNA and/or artificial oligonucleotides that can be modified or unmodified with high purity. Artificial oligonucleotides can act as RNA and DNA mimics but may have different sets of properties, such as higher affinities to their targets. These technologies have already enabled the use of synthetic oligonucleotides to construct synthetic biochemical circuits from simple components useful for the study of in-vitro transcriptional circuits. For example the use of two essential enzymes, bacteriophage T7 RNA polymerase and Escherichia coli ribonuclease H, together with synthetic oligonucleotides allowed the systematic construction of arbitrary circuits for the study of synthetic in-vitro transciption. In addition, scientists in recent years have shown that mathematical modeling can be used to guide the design process of this type of systems. Some of these experimental conditions yielded oscillating biochemical circuits. The notion here is that synthetic transcriptional oscillators could prove valuable for systematic exploration of biochemical circuit design principles. The understanding of these principles could allow scientist to design artificial cells and control nanoscale devices. Future studies will surely identify additional examples of direct communications between regulatory and general transcription factors and reveal how promoter groups and gene networks are regulated through these interactions. Finally, new insights into the specific biological processes in which they are involved will be gained.
What exactly is transcription?
Transcription refers to the transfer of genetic code information from one kind of nucleic acid to another. It refers to the process by which a base sequence of messenger RNA is synthesized by an RNA polymerase on a template of complementary DNA. On the other hand, the reversal of this process or flow of information is called “reverse transcription.” In reverse transcription the normal pattern is reversed, for example by a viral enzyme called “reverse transcriptase.” Furthermore, a polymerase associated with the process of transcription is called a “transcriptase”. The DNA-dependent RNA polymerase is an example for a transcriptase. The template is defined as “a single-stranded polynucleotide or the region of a polynucleotide that directs the synthesis of a complementary polynucleotide”. Experiments that aim to find out which portion of a DNA molecule is transcribed into RNA are called “transcript analysis”. Furthermore, the entire mRNA content of a cell or tissue is now called the “transcriptome”.
In bacteria, or prokaryotes, transcription is catalyzed by a single RNA polymerase. In e.coli the RNA polymerase is a large enzyme of almost 500 kilodaltons. The holoenzyme has five subunits, α2ββ’ω, and the core enzyme lacks the σ factor. An essential step in bacterial transcription is the binding of one of a number of dissociable accessory proteins, called the σ factor, to a core RNA polymerase to form a holoenzyme. In e. coli the core enzyme, α2ββ’ω with a mass of 379 kDa binds the σ70 factor to form the holoenzyme α2ββ’ω σ70, with a mass of 449 kDa. Only as a holoenzyme can RNA polymerase initiate transcription and it is thought that the holoenzyme binds weakly to the DNA and explores the DNA double helix until with the help of the σ factor it recognizes a promoter sequence to which it finely binds tightly. Finn et al. in 2000 determined the structures of the core RNA polymerase and the σ70 holoenzyme using cryo-electron microscopy and angular reconstruction.
Escherichia coli RNA polymerase (RNAP) is the most studied bacterial RNA polymerase and has been used as the model RNAP for screening and evaluating potential RNAP-targeting antibiotics. Murakami in 2013 reported the X-ray structure of the E. coli RNAP σ(70) holoenzyme which showed the σ region 1.1 (σ1.1) and the α subunit C-terminal domain in the context of an intact RNAP. The structure revealed that σ1.1 is positioned at the RNAP DNA-binding channel and completely blocks DNA entry to the RNAP active site. Furthermore, the σ1.1 contains a basic patch on its surface. It is thought that this batch may play an important role in DNA interaction to facilitate open promoter complex formation. In this structure the α-subunit C-terminal domain is positioned next to σ domain 4 with a fully stretched linker between the N- and C-terminal domains. The X-ray based model is depicted in the next figure.
Transcription involves RNA polymerases binding, initiation,elongation and termination.
During the initiation of the transcription cycle an RNA polymerase searches for a promoter site and binds to DNA, unwinds and separates the two strands. The separation occurs in such a way that one strand, the template strand, is copied, but not the other. Next, a base pair forms between a base in the template strand and a ribonucleotide triphosphate. The first nucleotide retains its three phosphate groups at the 5’ end as the RNA chain grows on the OH group attached to its 3’ carbon. During elongation the RNA strand grows from its 5’-end to its 3’ end, as the polymerase copies the template DNA strand from its 3’-end to its 5’-end. The RNA polymerase catalyzes the formation of a phosphodiester bond. The DNA duplex unwinds and a newly duplex is formed made of the newly synthesized RNA and DNA template strand, which extends for 10 to 12 bases behind the most recent DNA. Next, the growing RNA molecule detaches from the template and the DNA helix forms again. The new RNA strand has exactly the same sequence as the nontemplate strand of DNA, however, wherever DNA has a thymidine (T), the RNA has a uridine (U). During termination, the ending of chain growth, the RNA polymerase detaches from the DNA template and the growing RNA chain. Termination sites or sequences in the DNA signal the end of transcription. Many prokaryotic genes contain self-complementary sequences that can fold back on itself and form hairpin duplexes or stretches of U’s.
However, termination in eukaryotes is more complex and the elucidation of the exact mechanism or mechanisms is still under investigation. Termination can also occur at termination sites that do not have these features, in additon, for this to occur protein factors are needed. One of these termination assisting proteins is called rho (ρ). In addition, so called antiterminating proteins can allow elongation to continue through DNA sequences that otherwise can serve as termination sites.
Transcription of two genes.
RNA polymerase moves from the 3’ end of the template strand and creates an RNA strand that grows in a 5’ to 3’ direction because it must be antiparallel to the template strand. Some genes are transcribed from one strand of the DNA double helix whereas other genes are transcribed from the other as the template. Furthermore, a uracil is being added to the 3’ end of the transcript for gene 1. Querying the Pubmed structure data base revealed that many solved structures are available for polymerase complexes indicating the importance of these protein complexes.
Zhang et al. in 2012 have determined crystal structures at 2.9 and 3.0 Å resolution of functional transcription initiation complexes comprising Thermus thermophilus RNA polymerase, sigma factor A (σ), and a promoter DNA fragment corresponding to the transcription bubble and downstream double-stranded DNA of the RNAP-promoter open complex. The structures showed that σ recognizes the –10 element and discriminator element through interactions that include the unstacking and insertion into pockets of three DNA bases and that RNA polymerase recognizes the –4/+2 region through interactions that include the unstacking and insertion into a pocket of the +2 base. Furthermore the structures also revealed that interactions between σ and template-strand single-stranded DNA (ssDNA) preorganize the template-strand ssDNA to engage the active center of RNA polymerase. The model of the crystal structure of the bacterial RNA polymerase (RNAP)-promoter complex (RPo) in complex with a ribonucleotide primer (RPo-GpA) is shown below.
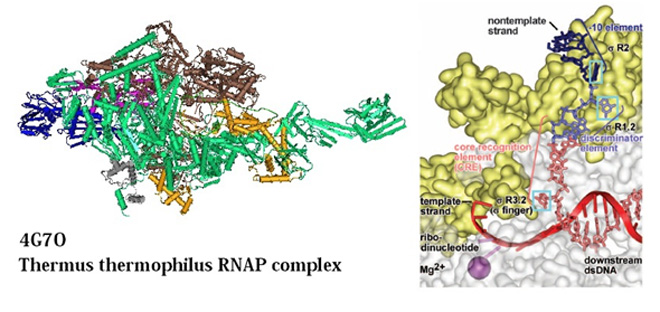
Model of the bacterial RNA polymerase (RNAP)-promoter complex (RPo) in complex with a ribonucleotide primer (RPo-GpA).
The crystal structure of a bacterial RNA polymerase (RNAP)-promoter complex (RPo) in complex with a ribonucleotide primer (RPo-GpA) is illustrated at the left part of the panel. The interactions of RNAP and the σ factor with the transcription-bubble non-template strand, transcription-bubble template strand, and the downstream dsDNA is shown at the right part of the panel. (Source: Zhang et al. 2012). The sigma factor (σ factor) is a protein needed for the initiation of RNA synthesis. This bacterial transcription initiation factor enables specific binding of RNA polymerase to gene promoters. However, each specific sigma factor used to initiate transcription of a given gene will vary depending on the gene and on the environmental signals needed to initiate transcription of that gene. In addition, every molecule of RNA polymerase holoenzyme contains exactly one sigma factor subunit.
Bye 1 is a transcription factor that links histones to post-translational modification events.
Kinkelin et al. in 2013 reported crystal structures of the nuclear protein bypass of Ess1 (Bye1) TFIIS-like domain (TLD) bound to Pol II and three different polymerase II-nucleic acid complexes. The researchers could show that like TFIIS, Bye1 binds with its TLD to the polymerase II jaw and funnel. Furthermore, it was demonstrated that Bye1 is recruited in vivo to chromatin via its TLD and occupies the 5'-region of active genes. The paper showed that a plant homeo domain (PHD) in Bye1 binds histone H3 tails with trimethylated lysine 4. This interaction is enhanced by the presence of neighboring posttranslational modifications (PTMs) that mark active transcription and is impaired by repressive PTMs. The scientists identified putative human homologs of Bye1, the proteins PHD finger protein 3 and death-inducer obliterator. Both proteins are implicated in cancer. These results establish Bye1 as a chromatin transcription factor that links histones with active PTMs to transcribing Polymerase II. Bypass of Ess1 (Bye1) is a nuclear protein with a domain resembling the central domain in the transcription elongation factor TFIIS.
What allowed the scientists to establish their model?
The researchers used purified cloned proteins together with Surface Plasmon Resonance, crystallization of the complexes followed by determining the X-ray structure, chromatin fractionation, histone peptide microarrays, synthetic lethality screening, in vitro transcription assays, RNA extension assays, chromatin immune precipitation (ChIP) and gene averaged profiling. A model of this complex is shown next.
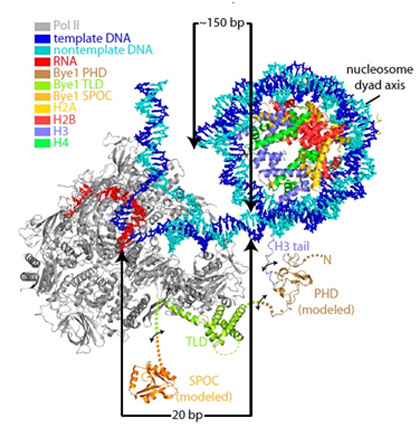 |
Model of a polymerase II-Bye 1-nucleosome complex.
This model shows that Bye associates with active genes in front of the +2 nucleosome. The model is based on crystal structures and ChIP occupancy peak positions (Source: Kinkelin et al., 2013).
|
The central dogma of molecular biology first stated by Francis Crick in 1958 and re-stated in a Nature paper published in 1970 describes the “information flow in biological systems”. It deals with the detailed residue-by-residue transfer of sequential information. It states that such information cannot be transferred back from protein to either protein or nucleic acid and has been also described as "DNA makes RNA makes protein." Furthermore, since it is a simplification it does not make it clear that the sequence hypothesis as stated by Crick does not preclude the reverse flow of information from RNA to DNA, but only the reverse flow from protein to RNA or DNA.
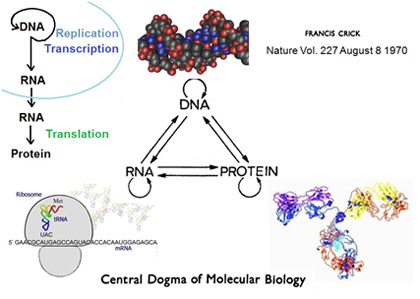 |
This graphics (left) illustrates the central dogma and its expansion as new knowledge about the nature of different types of RNAs became available.
The flow of information and substances in a cell is even more complex. This is illustrated in more detail next. Regulatory feedback loops are depicted as well.
|
The exponential increase in RNA research has in recent years tremendously increased our knowledge about their function. This has led to our expanded understanding of complex regulatory pathways implied by the central dogma. The next graphic shows the current view how RNA processing is thought to occur in a cell.
Eukaryotes express many functional non-protein-coding RNAs (ncRNAs) that used to be thought of as “junk DNA’ or the “dark matter” of the DNA. These RNAs participate in the processing and regulation of other RNA molecules common patterns have emerged that form a network-like RNA infrastructure. For more details see Collins & Penny in Trend in Genetics, 2009.
How can we study RNA transcripts of genes?
Over the years scientists have devised a variety of techniques to study RNA transcripts. Some of them detect the presence of a transcript and give some information of its length whereas others enable the start and end of the transcript to be mapped and the position of introns to be located. The following is a list of these methods:
Northern hybridization
RNA molecules present in RNA extracts are separated by electrophoresis, for example, in an agarose gel, using denaturing buffers to ensure that the RNAs do not form inter- or intramolecular base pairs. After electrophoresis, the gel is blotted onto a nylon, a nitrocellulose or a polyvinylidene fluoride (PVDF) membrane followed by hybridization with a labeled probe. If the probe is a cloned gene, the band that appears on the color or radioactively developed membrane is the transcript of the gene. The size can be determined from its position within the gel in relation to RNA marker molecules or RNA isolated from different tissues that are run in different lanes of the gel. This allows to find out if the gene is differentially expressed.
Transcript mapping by hybridization between gene and RNA
DNA-mRNA hybridization can be used to investigate if incomplete or complete cDNA synthesis occurred. If a hybrid is formed between a DNA strand that contains a gene and its mRNA the boundaries between double- and single-stranded regions will mark the start and end points of the mRNA. Introns that are present in the DNA but not in the mRNA will “loop out” as additional single-stranded regions. S1 nuclease degrades single-stranded DNA or RNA polynucleotides including the single-stranded ends of predominantly double-stranded molecules. However S1 nuclease has no effect on double-stranded DNA or on DNA-RNA hybrids. Singel-stranded DNA fragments protected from S1 nuclease digestion can be recovered by treatment with alkali. S1 nuclease mapping allows the localization of the starting-point and end-point of a transcript.
Transcript analysis by primer extension
Primer extension can only be used if at least part of the sequence of a transcript is known. In this technique a short oligonucleotide primer must be annealed to the RNA at a known position. Usually the primer anneals within 100-200 nucleotides of the 5’ end of the transcript and is extended by reverse transcriptase which is a cDNA reaction. The 3’ end of the newly synthesized strand of DNA corresponds with the 5’ terminal end of the transcript. Determination of the length of the single-stranded DNA molecule and correlating this information with the annealing position of the primers allows location of the position of this terminus.
Transcript analysis by PCR
A modified method of a standard reverse trancriptase PCR procedure called rapid amplification of cDNA ends (RACE) can be used to identify the 5’ and 3’ termini of RNA molecules and allow to map the ends of a transcript. Several RACE methods have been developed over the years.
Digital PCR can be used to determine the number of transcripts
Farago et al. in 2003 have shown that digital PCR can be used to determine the number of transcripts from single neurons after patch-clamp recoding.
How can the yields of full-length RNA from an in-vitro transcription reaction be maximized?
The quality and quantity of RNA produced in an in-vitro transcription reaction is dependent upon several factors. The size of the RNA transcript, template concentration, reaction time and temperature, all influence the yield of the final full-length transcript. During an individual application, the starting template and the resulting transcript determine how a reaction needs to be modified to increase the final yields. In general, T7, T3, or SP6 RNA polymerase are used for in-vitro transcription reactions. Each polymerase recognizes a short, well-defined, phage promoter sequence with a high degree of specificity that has a minimal length of 20 nucleotide bases. In-vitro transcription reactions usually include ribonucleoside tri-phosphates (rNTPs) at concentrations of 0.5 mM per each nucleotide, reaction buffer, a linear DNA template, amounts of 1 to 2 µg, and the appropriate bacteriophage RNA polymerase. On average, these reactions produce 10 to 40 µg of RNA. However, since increasingly techniques such as gene expression profiling using microarrays, RNA interference (RNAi) gene-silencing, in-vitro translation, ribozymes, and RNA structure studies require larger quantities of RNA commercial in-vitro transcription kits have been developed to address this need. Starting with 1 µg of template up to 100 or 200 µg of RNA may be produced. Even though reaction conditions in commercial kits have been optimized to maximize the RNA yield from control templates which typically can produce 1 to 2 kB transcripts, each unique transcript may need adjustments to the protocol to maximize yields.
Template preparation
The DNA template used should be linear, double-stranded, and relatively clean. Typically templates used include linearized plasmids that contain blunt or 5’-protruding ends, PCR products or cDNAs. The templates should be free of RNase and other contaminations such as phenol, trace metals and sodium dodecyl sulfate (SDS). The treatment of the DNA with proteinase K, followed by phenol-chloroform extraction and ethanol precipitation usually allows producing a sufficiently clean template. The analysis of the template with the help of an agarose or polyacrylamide gel and staining with ethidium bromide or syber green allows verifying that the template is pure enough.
Maximizing yield for long RNA transcripts
Template concentration and reaction time: For the production of a long-transcript a higher concentration of template DNA is less critical. Sometimes increasing the amount of template reduces the total reaction time to yield the RNA transcript.
Optimizing long transcripts from limited amounts of template
If the amounts of template DNA is limited to less than 1 µg the reaction time and/or temperature may need to be increased, or the reaction may need to be scaled up, to maximize yields. Reaction incubation times may need to be increased to 2 to 3 hours.
Reaction temperature for a long transcript
Increasing the reaction temperature from 37 °C to 42 °C will increase the rate at which the RNA is produced and also increases the maximal yield that can be attained. This temperature effect is more significant for lower template concentrations. A limited amount of template may demand that the reaction is incubated at 42 °C for 3 to 4 hours.
Scale-up for long transcripts
To achieve even higher yields all components of the reaction mixture including the template may need to be scaled up by 6 to 10 fold. Because the initiation of the reaction is the rate-limiting step in a transcription reaction, the reaction dynamics in a short-transcript reaction differ from those in long-transcript reaction. The best way to optimize transcription of short RNAs is to maximize the number of possible transcription initiations. This can be achieved by increasing template concentration or reaction times.
Template size and concentration for a short transcript
The increase of the template concentration in an in-vitro transcription reaction will result in an increase of the yield of a short RNA transcript. Usually in-vitro transcription reactions use 1 µg of template. However, if a 75 base pair (bp) double-stranded oligonucleotide template is used 20 picomoles of DNA will be used. On the other hand, if 1 µg of a 4.2 kilo base (kb) linear plasmid is used only 360 femtomoles of DNA is present. Therefore, 1 µg of an oligonucleotide template produces more short RNA than 1 µg of a plasmid template if all other parameters are equal in the reaction.
Reaction time for a short transcript
Increasing the reaction time will increase the number of transcription initiation events and significantly increase the yield of the RNA. However, after a reaction time of 3 to 4 hours the yield in general will plateau.
Reaction temperature for a short transcript
Typically in-vitro transcription reactions are performed at 37o°C. The increase of the temperature to 42 °C can increase the yields and shorten reaction times.
In conclution, RNA yields for both long and short transcripts from in-vitro transcription reactions can be maximized by adjusting the template DNA concentration, the reaction time, and the reaction temperature.
Template preparation
Transcription templates can either be synthetic DNA, PCR products, or linearized vectors. Synthetic templates can be purified using reversed phase HPLC, ion exchange chromatography, an oligonucleotide purification cartridge, desalted by butanol precipitation or ethanol precipitation, or purified by Urea-PAGE. Furthermore, synthetic templates can be used for the antisense strand (cDNA).
References
Brown, T.A.; Gene cloning and DNA analysis. An introduction. Sixth edition. Wiley-Blackwell. 2010.
Crick, F.H.C. (1958): On Protein Synthesis. Symp. Soc. Exp. Biol. XII, 139-163.
Crick, F (August 1970). "Central dogma of molecular biology". Nature 227 (5258): 561–3. Bibcode:1970 Nature.227..561C. doi:10.1038/227561a0. PMID 4913914.}
Collins LJ, Penny D.;The RNA infrastructure: dark matter of the eukaryotic cell? Trends Genet. 2009 Mar;25(3):120-8. doi: 10.1016/j.tig.2008.12.003. Epub 2009 Jan 24.
Nóra Faragó, Ágnes K. Kocsis, Sándor Lovas, Gábor Molnár, Eszter Boldog, Márton Rózsa, Viktor Szemenyei, Enikő Vámos, Lajos I. Nagy, Gábor Tamás, and László G. Puskás; Digital PCR to determine the number of transcripts from single neurons after patch-clamp recording. BioTechniques 54:327-336 ( June 2013) doi 10.2144/000114029.
Robert D. Finn, Elena V. Orlova, Brent Gowen, Martin Buck, and Marin van Heel; Escherichia coli RNA polymerase core and holoenzyme structures. The EMBO Journal vol. 19 No. 24 pp. 6833-6844, 2000.
Magali Frugier, Catherine Florentz, Mir Wais Hosseini, Jean-Marie Lehn and Richard Giegd; Synthetic polyamines stimulate in-vitro transcription by T7 RNA polymerase2784-2790 Nucleic Acids Research, 1994, Vol. 22, No. 14.
Kinkelin, K., Wozniak, G.G., Rothbart, S.B., Lidschreiber, M., Strahl, B.D., Cramer, P.; Structures of RNA polymerase II complexes with Bye1, a chromatin-binding PHF3/DIDO homologue. (2013) Proc.Natl.Acad.Sci.USA 110: 15277.
Jongmin Kim and Erik Winfree; Synthetic in-vitro transcriptional oscillators. Molecular Systems Biology 7:465
Lewin’s Genes I to XI. Oxford University Press.
Lodish, Harvey; Molecular Cell Biology – 2nd to 7th edition. Scientific American Books.
Craig T. Martin, Daniel K. Muller, and Joseph E. Coleman; Processivity in Early Stages of Transcription by T7 RNA Polymerase. Biochemistry 1988, 27, 3966-3974.
Murakami KS; X-ray crystal structure of escherichia coli RNA polymerase sigma70 holoenzyme. J.Biol.Chem. (2013) 288 p.9126.
Patrushev LI, Bocharova TN, Khesin RB.;The effect of various templates and oligonucleotide primers on RNA and poly (A) synthesis by E. coli and T7 RNA polymerases. FEBS Lett. 1978 Feb 1;86(1):108-12.
Yu Zhang, Yu Feng, Sujoy Chatterjee, Steve Tuske, Mary X. Ho, Eddy Arnold, Richard H. Ebright; Structural Basis of Transcription Initiation. Science 338, 1076 (2012).
---...---
10/28/2013