Isoelectric and Isoionic pH, the key to isoelectric focusing
Isoelectric focusing (IEF), or electrofocusing, is a biochemical technique that allows the separation of bio-molecules such as proteins and peptides according to their different isoelectric points (pIs). This type of electrophoresis uses polyacrylamide gels to separate proteins and peptides in electrophoretic zones. The technique is made possible by the fact that the overall charge on the molecule of interest is a function of the pH of its surrounding environment. The selection of the buffer system, in this case combinations of ampholytic buffers allows the development of an isoelectric gradient that can span over several pH ranges.
Peptides and proteins are polyprotic systems which have an isoelectric or isoionic point (the pI). The isoionic point or isoionic pH is obtained when a pure, neutral polyprotic acid is dissolved in water. The isoelectric point is the pH at which the average charge of the polyprotic acid is zero (0). In solution, most of the molecules will be in the uncharged form HA, and the concentrations of the acid H2A+ and the base A- will be in equilibrium with HA.
To explain these phenomena in more detail we will have to review a few basic biochemical concepts first. If two types of ions that can chemically interact with each other and are present in different quantities come into contact with each other they can establish a chemical equilibrium. Next, let us review the chemical equilibrium in aqueous solutions. The concept of chemical equilibrium provides the foundation for chemical calculations useful to chemical analysis and other areas of science such as biochemistry, biology, medicine, geology, oceanography and other scientific fields.
For a reaction aA + bB ⇌cC + dD we can write the equilibrium constant, K,
in the form Equilibrium constant K = [C]c[D]d / [A]a[B]b
And the equilibrium constant for the reaction H2O ⇌H+ + OH- is called Kw (= [H+][OH-) and has the value 1.0x 10-14 at 25 C.
This equation describes the autoproteolysis constant of water and is called Kw. That means that water can dissociate into protons, H+, and hydroxyl anions, OH-. The concentration of H+ and OH- are both 1.0 x 10-7 M in pure water. Remember, the mole is a unit of measurement used in chemistry to express amounts of a chemical substance. It is defined as an amount of a substance that contains as many elementary entities (e.g., atoms, molecules, ions, electrons) as there are atoms in 12 grams of pure carbon-12 (12C), the isotope of carbon with atomic weight 12. This corresponds to a value of 6.02214179(30) × 1023 elementary entities of the substance. This is one of the base units in the “International System of Units”, and has the unit symbol mol.
Let us review the concept of the pH next. An approximate definition of pH is the negative logarithm of the H+ concentration.
Approximate definition of pH:pH = - log[H+]
The pH can be defined more accurately in terms of activities but for most purposes this equation is a good working definition. Please review books for “Quantitative Chemical Analysis” if you want to study this subject in more detail. To explain the concept of the pI more clearly we still need to review a few more basic definitions such as the concept of acid and bases, monoprotic and polyprotoic acids, pKa and pKb.
An acid (from the Latin acidus/acēre or sour) is defined as a substance that reacts with a base. Acids can be identified as tasting sour, reacting with metals such as calcium, and bases like sodium carbonate. Aqueous solutions of acids have a pH below 7. The more acidic the acid is the lower the pH.
In the presence of water an acid, denoted HA, can react with water molecules by donating a proton to the water molecule: Dissociation of an acid (in this case a week acid) can be expressed as:
A + H2O ⇌H3O+ + A-, and HA ⇌H+ + A-, or as a dissociation constant
Ka = [H+][A-] / [HA] which is called the acid dissociation constant.
Similarly, the equilibrium constant for bases is called the base hydrolysis constant or Kb or sometimes, mistakenly, called the base “dissociation’ constant.
BA + H2O ⇌ BH+ + OH-, and base hydrolysis constant
Kb = [BH+][OH-] / [B]
Monoprotic and polyprotoic acids: Monoprotic acids are acids that can donate one proton per molecule during dissociation or ionization as shown below:
HA(aqueous) + H2O(liquid) ⇌H3O+(aq) + A−(aq) Ka
Some examples of monoprotic acids include hydrochloric acid (HCl), nitric acid (HNO3), and organic acids such as formic acid (HCOOH), acetic acid (CH3COOH), and benzoic acid (C6H5COOH).
Polyprotic acids, also known as polybasic acids, are able to donate more than one proton per molecule. Diprotic acids can potentially donate two protons and triprotic acids can donate three protons. A diprotic acid (H2A) can dissociate in two steps depending on the pH. Each dissociation step has its own dissociation constant, Ka1 and Ka2.
H2A(aq) + H2O(l) ⇌ H3O+(aq) + HA−(aq) Ka1
HA−(aq) + H2O(l) ⇌ H3O+(aq) + A2−(aq) Ka2
Typically the first dissociation constant is greater than the second; i.e., Ka1 > Ka2. Sulfuric acid (H2SO4) can donate one proton to form a bisulfate anion (HSO4−), with a large Ka1; next it can donate a second proton to form a sulfate anion (SO42-), with an intermediate strength Ka2. The large Ka1 for the first dissociation makes sulfuric a strong acid. Similarly, the weak and unstable carbonic acid (H2CO3) can lose one proton to form a bicarbonate anion (HCO3−) and lose a second to form a carbonate anion (CO32-). Both Ka values are small, with Ka1 > Ka2 . Triprotic acids (H3A) can undergo one, two, or three dissociations and have three dissociation constants, where Ka1 > Ka2 > Ka3.
H3A(aq) + H2O(l)⇌H3O+(aq) + H2A−(aq) Ka1
H2A−(aq) + H2O(l) ⇌ H3O+(aq) + HA2−(aq) Ka2
HA2−(aq) + H2O(l) ⇌ H3O+(aq) + A3−(aq) Ka3
Let us calculate the isoelectric point for an amino acid. We selected histidine as an example. Histidine has the following reported pKas: pKa1 = 1.77, pKa2 = 6.10 and pKa3 =9.18. The histidine molecule can have different net charges depending on the surrounding pH which is illustrated with chemical structures shown below.
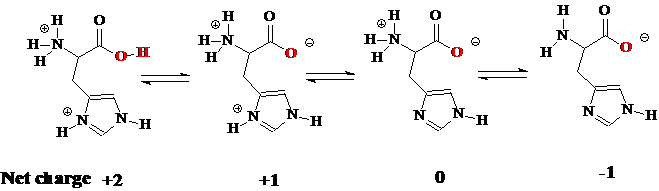
At the pI the net charge of the molecule is zero. To find the pI we need to average the two pKa values on either side of the neutral form of the amino acid. For histidine it can be calculated as follows:
(pKa2 + pKa3)/2 = pI => (6.10 + 9.18)/2 = 7.64
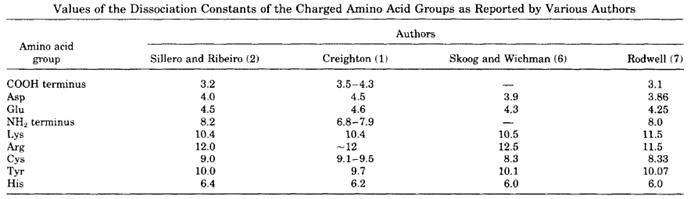
And so on for each selected amino acid. The following table lists pKa values for the amino acids naturally occurring in proteins.
Amino Acid pKa Values
Note: The measurement and calculation of the pKa is not always very accurate and different authors have reported slightly different values in the past. The table shown below taken from Patrickios and Yamasaki (1995) illustrates this. However, Patrikios and Yamasaki report that theoretical isoelectric points determined by using more complex computational procedures are on average in good agreement with the experimental values.
In a protein molecule
ai the number of negatively charged groups of type
i depend on the total number of acidic groups
i,
Ai, the
pH, and the dissociation constant of the particular acidic group,
pKAi.
ai = Ai/[1 + 10(pKAi- pH)]
Similarly, the number of positively charged groups can be calculated accordingly.
bi = Bi/[1 + 10(pH - pKBi)]
At the pI of a protein the total number of negatively charged groups should be equal to the total number of positively charged groups. This can be expressed as follows:
Σi ai = Σi bi at pH = pI
Therefore, to calculate the total pI of a protein the following equation can be used:
Σi Ai/[1 + 10(pKAi- pI)] = Σi Bi/[1 + 10(pI - pKBi)]
Computational procedures to calculate this have been developed and many of these are available on the web.
http://www.biosyn.com/PeptidePropertyCalculator/PeptidePropertyCalculator.aspx
http://web.expasy.org/compute_pi/
http://isoelectric.ovh.org/
http://www.geneinfinity.org/sp/sp_proteinpinmw.html
and others.
The next table shows a list of experimental and calculated isoelectric points of proteins published by Patrickios and Yamasaki in 1995. These values can be used to design purification and separation protocols for protein analysis method.
Experimental and Calculated Isoelectric Points of Proteins
 References
References
Harris, Daniel; Quantitative Chemical Analysis. 5th Edition 1999. W. H. Freeman and Company.
Patrickios CS, Yamasaki EN,;Anal Biochem. Polypeptide amino acid composition and isoelectric point. II. Comparison between experiment and theory. 1995 Oct 10;231(1):82-91.