The degradation of DNA and RNA, which hinders their potential as therapeutic agents, is primarily due to the action of nucleases. These enzymes, including endonuclease and exonuclease, are involved in various intracellular processes such as DNA repair, replication, recombination, and anti-viral defense. They target DNA and RNA, leading to their rapid degradation. The RNA interference mechanism can also contribute to the degradation of double-stranded RNA or mRNA (Bernstein et al., 2001).
In recent decades, scientists have developed xeno nucleic acids (XNAs), representing artificially modified nucleic acid analogs to improve natural nucleic acids.
However, modifications of the base moiety and the phosphodiester backbone may render the complementary base pairing ability and nuclease resistance; modification of the sugar group may affect both properties.
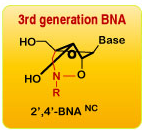
One type of XNAs includes 2’-modified analogs, such as 2’-deoxy-2’-fluoroarabino nucleic acid (FANA) and 2’-O-methoxyethyl-RNA (MOE). Bridged nucleic acid (BNA) represents another category comprised of conformation-locked analogs such as locked nucleic acid (LNA) with a bridge linking 2’-O and 4’-C of the ribose moiety. 2'-O,4'-aminoethylene bridged nucleic acid (2',4'-BNANC) is the third-generation BNA containing a six-member bridged structure with an N-O linkage. An increased conformational inflexibility of ribose renders greater binding affinity to complementary single-stranded RNA or double-stranded DNA. Scientists also developed XNAs with a 6-membered ring sugar group, for example, 3’-fluoro hexitol nucleic acid (FHNA) (Morihiro et al., 2017).
 https://www.biosyn.com/bna-synthesis-bridged-nucleic-acid.aspx
https://www.biosyn.com/bna-synthesis-bridged-nucleic-acid.aspx
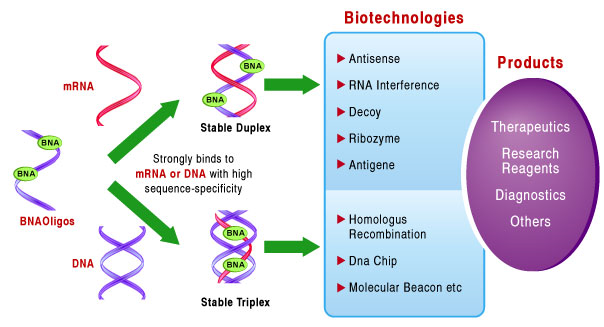
The emergence of Xeno Nucleic Acids (XNAs) presents a promising avenue for using oligonucleotides in therapy, with potential applications that surpass their unmodified precursors. With enhanced resistance to nucleases, heightened specificity, and increased base-pairing potential, a multitude of clinical applications are on the horizon. These include antisense LNA oligodeoxynucleotides that bind to complementary H-Ras mRNA or miR-17-5p microRNA (miRNA) to inhibit cancer progression (Fluiter et al., 2005; Jin et al, 2015). A gapmer incorporating BNANC efficiently degraded CUG expanded repeat RNA, which causes myotonic dystrophy (Manning et al., 2017). For Duchenne muscular dystrophy, splice-switching oligonucleotides with 2′-O-MOE was effective in modulating the pre-messenger RNA splicing of dystrophin (Yang et al, 2013). For RNA interference, the LNA-based siRNA therapeutics exhibited greater functionality/stability (Elmén et al., 2005).
Aptamers consist of single-stranded RNA or DNA oligonucleotides whose 3-dimensional structure has been exploited to isolate species that bind specifically to therapeutic targets. The RNA aptamer Pegaptanib targeting vascular endothelial growth factor (VEGF) was the first to be approved by the FDA for age-related macular degeneration (Lee et al., 2015). To identify, a library consisting of randomly sequenced DNA or RNA oligonucleotides is screened for binding to a target ligand. The bound oligonucleotides are recovered and amplified by PCR before repeating the selection cycle to enrich the specifically bound sequences. The isolated oligonucleotides must be reverse-transcribed to identify an RNA aptamer before amplifying.
Incorporating XNAs like BNA to render nuclease resistance can greatly improve the stability of aptamers.
However, most modified nucleosides are poor substrates for polymerases and reverse transcriptase. One solution is to introduce XNAs after identifying the aptamers. Another approach is to generate polymerase mutants capable of recognizing XNA-based aptamers. Using Tgo DNA polymerase derived from Thermococcus gorgonarius, PolC7, Pinheiro et al. generated a variant capable of reverse transcribing LNA-based oligonucleotides (Pinheiro et al., 2012), which the research group used to isolate a HNA-based aptamer binding to HIV’s trans-activating response RNA (TAR). Another group reported that LNA-based oligonucleotides could be reverse transcribed using SuperScript III Reverse Transcriptase (Crouzier et al., 2012). A polymerase mutant capable of amplifying 2’-OMe containing oligonucleotide was also generated (Chen et al 2016).
Bio-Synthesis, Inc. has been at the forefront of oligonucleotide technologies for 35 years. Ever since the introduction of solid phase nucleotide synthesis, it has kept pace with the latest breakthroughs in the nucleotide modification chemistry. Its recent acquisition of a license from BNA Inc. of Osaka, Japan, for the manufacturing and distribution of BNANC, a third generation of BNA oligonucleotides, is in keeping with this commitment. In recent days, the company has expanded its operation to align closely with the developments in therapy. To meet the demands of therapeutic application, its oligonucleotide products are approaching GMP grade. Bio-Synthesis, Inc. has recently entered into collaborative agreement with Bound Therapeutics LLC. to synthesize miR-21 blocker using BNA for developing potential triple-negative breast cancer treatment. The BNA technology that we offer provides superior, unequaled advantages in base stacking, binding affinity, aqueous solubility, and nuclease resistance. It also improves the formation of duplexes and triplexes by reducing the repulsion between the negatively charged phosphates of the oligonucleotide backbone. Its single-mismatch discriminating power was especially useful for diagnosis (for example, FISH using DNA probes). More importantly, BNA oligonucleotide exhibits lesser toxicity than other modified nucleotides for clinical application.
References
Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. (2001). Nature. 409, 363–366. PMID 1120174 . doi:10.1038/35053110.
Chen T, Hongdilokkul N, Liu Z, Adhikary R, Tsuen SS, Romesberg FE. Evolution of thermophilic DNA polymerases for the recognition and amplification of C2'-modified DNA. (2016) Nat Chem. 8:556-62 PMID: 27219699 PMCID: PMC4880425 DOI: 10.1038/nchem.2493
Crouzier L, Dubois C, Edwards SL, Lauridsen LH, Wengel J, Veedu RN. Efficient Reverse Transcription Using Locked Nucleic Acid Nucleotides towards the Evolution of Nuclease Resistant RNA Aptamers. (2012). PLoS One. 2012; 7: e35990. PMCID: PMC3338489 PMID: 22558297 doi: 10.1371/journal.pone.0035990
Elmén J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Ørum H, Koch T, Wahlestedt C. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. (2005). Nucleic Acids Res. 33:439-47. PMID: 15653644
Fluiter K1, Frieden M, Vreijling J, Rosenbohm C, De Wissel MB, Christensen SM, Koch T, Ørum H, Baas F. On the in vitro and in vivo properties of four locked nucleic acid nucleotides incorporated into an anti-H-Ras antisense oligonucleotide. (2005). Chembiochem. 6:1104-9. PMID: 24244378 PMCID: PMC3824000 DOI: 10.1371/journal.pone.0078863
Manning KS, Rao AN, Castro M, Cooper TA. BNANC Gapmers Revert Splicing and Reduce RNA Foci with Low Toxicity in Myotonic Dystrophy Cells. (2017). ACS Chem Biol. 12:2503-2509. PMID: 28853853 PMCID: PMC5694563 doi: 10.1021/acschembio.7b00416.
Morihiro K, Kasahara Y, Obika S. Biological applications of xeno nucleic acids. (2017). Mol Biosyst. 13:235-245. PMID: 27827481 doi: 10.1039/c6mb00538a.
Lee JH, Canny MD, De Erkenez A, Krilleke D, Ng YS, Shima DT, Pardi A, Jucker F. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. (2005). Proc Natl Acad Sci U S A. 102:18902-7.
Pinheiro VB1, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, Chaput JC, Wengel J, Peak-Chew SY, McLaughlin SH, Herdewijn P, Holliger P. Synthetic genetic polymers capable of heredity and evolution. (2012) Science. 36: 341–344. PMCID: PMC3362463 PMID: 22517858 doi: 10.1126/science.1217622
Yang L, Niu H, Gao X, Wang Q, Han G, Cao L, Cai C, Weiler J, Yin H. Effective exon skipping and dystrophin restoration by 2′-O-Methoxyethyl antisense oligonucleotide in dystrophin-deficient mice. (2013). PLoS One. 8: e61584. PMCID: PMC3637291 PMID: 23658612 doi: 10.1371/journal.pone.0061584