Enhancer RNAs add a crucial regulatory layer to the genome. Kim et al. (2010) identified a new class of RNAs called enhancer RNAs or eRNAs. Enhancers are cis-regulatory gene elements and are crucial for controlling temporal and cell-type specific patterns of gene expression.
Several classes of non-coding RNAs (ncRNAs), including microRNAs (miRNAs) and long non-coding RNAs (ncRNAs), play diverse roles in post-transcriptional regulation of mRNA stability and epigenetic control of chromatin activities.
Enhancer RNA or eRNA are RNAs transcribed by RNA polymerase II (RNAPII) from the domain of transcription enhancers. They are a class of short, non-coding RNAs transcribed from DNA enhancer regions, 50 to 2,000 nucleotides in length. Enhancer regions are bidirectionally transcribed into enhancer RNAs (eRNAs). As a response to signaling pathways, intergenic DNA elements known as enhancers regulate target gene transcription. Enhancers interact with promotors over large genomic distances. Enhancers contain binding sites for transcription factors promoting RNA polymerase II (RNAPII) recruitment and activation. Enhancers carry unique epigenetic marks, distinguishing them from promoters.
These regulatory elements also have an open chromatin conformation that increases accessibility to transcription factors and RNAPII. Rahman et al. recently reported that eRNAs are localized exclusively in the nucleus. The induction of eRNAs occurs with similar kinetics as that of target mRNAs. eRNAs are beginning to develop mostly as enhancers. The steady-state levels of eRNAs remain lower than those of their related mRNAs and RNAs at the single-allele level. Also, eRNAs are rarely co-expressed with their target loci. However, active gene transcription does not require continuous transcription of eRNAs or the accumulation of eRNAs at enhancers.
Recently developed genome-wide sequencing methods allow studying stimulus-dependent enhancer functions in tissue cells. Kim et al. (2010) found that the level of eRNA expression at neuronal enhancers positively correlates with the level of mRNA synthesis at nearby genes. These observations suggest that eRNA synthesis occurs specifically at enhancers that actively promote mRNA synthesis and that a widespread mechanism of enhancer activation involves RNAPII binding and eRNA synthesis.
Kim et al. suggested that establishing and maintaining the chromatin landscape at enhancers requires eRNA synthesis for enhancer function. It is also possible that the eRNA transcripts are functionally crucial by themselves. A typical gene transcribed by RNA polymerase II has a promotor that extends upstream from the site where transcription is initiated (Figure 1).
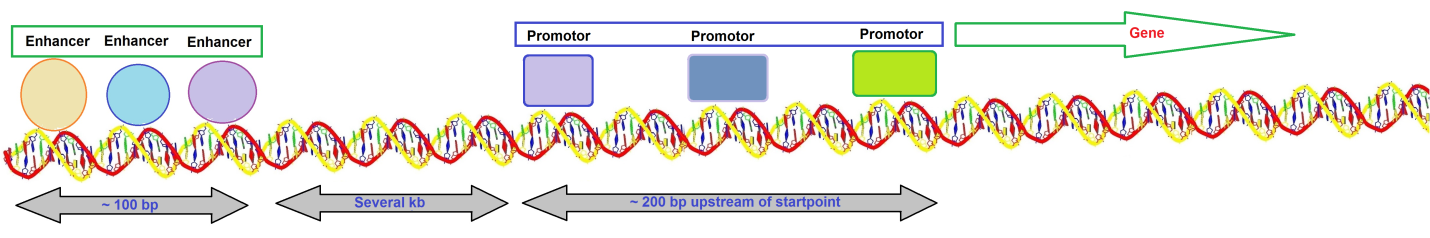
Figure 1: Overview of a typical gene transcribed by RNA polymerase II.
The promotor contains several short sequence elements that bind transcription factors, <10 base pairs (bp) in length, and promotors are dispersed over a sequence region >200 bp. Enhancers contain a more closely packed array of elements that bind transcription factors. Enhancer regions appear to be located at a distance of several kilobases (kb). The DNA duplex may be coiled or rearranged such that transcription factors at the promotor and the enhancer interact to form a large protein-DNA complex.
Enhancers contain bidirectional elements that allow assisting initiation. The presence of a promotor increases its activity. The enhancer is located distinct from the promotor, and its position relative to the promotor can vary substantially. An enhancer can stimulate any promotor placed in its vicinity. Enhancers often show redundancy in function. DNA must be able to form a loop structure if proteins bound at an enhancer several kb distant from a promotor interact directly with proteins bound in the vicinity of the starting point such that the enhancer and promotor are near to each other. Enhancers may function by bringing proteins into the area of the promotor.
Methods that allow analysis of eRNAs:
- Reverse transcription-PCR (RT-PCR)
- RNA fluorescence in situ hybridization (RNA-FISH)
- RNA polymerase II chromatin immunoprecipitation coupled with high-throughput sequencing (RNAPII ChIP–seq)
- Global run-on sequencing (GRO-seq)
- 5′GRO-seq or GRO-cap
- BruUV-seq
- RNA-seq (total)
- RNA-seq (poly(A))
- Cap analysis of gene expression (CAGE) followed by deep sequencing
- Chromatin- bound RNA-seq
- RNA-Seq in isolated 'transcription factories'
- Native elongating transcript sequencing (NET-seq)
- RNA capture sequencing (CaptureSeq)
- Chromatin isolation by RNA purification (ChIRP-seq)
Reference
Bose DA, Berger SL. eRNA binding produces tailored CBP activity profiles to regulate gene expression. RNA Biol. 2017 Dec 2;14(12):1655-1659. [PMC]
Jourdain AA, Koppen M, Rodley CD, Maundrell K, Gueguen N, Reynier P, Guaras AM, Enriquez JA, Anderson P, Simarro M, Martinou JC. A mitochondria-specific isoform of FASTK is present in mitochondrial RNA granules and regulates gene expression and function. Cell Rep. 2015 Feb 24;10(7):1110-21. [Cell Reports]
Kim, T.-K., Hemberg, M., Gray, J. M., Costa, A. M., Bear, D. M., Wu, J., … Greenberg, M. E. (2010). Widespread transcription at neuronal activity-regulated enhancers. Nature, 465(7295), 182–187. [Nature]
Lewin, Benjamin; Genes VII chapter 20, 2000.
Li, W., Notani, D. & Rosenfeld, M. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet 17, 207–223 (2016). [Nature Reviews genetics]
Rahman, S., Zorca, C. E., Traboulsi, T., Noutahi, E., Krause, M. R., Mader, S., & Zenklusen, D. (2017). Single-cell profiling reveals that eRNA accumulation at enhancer–promoter loops is not required to sustain transcription. Nucleic Acids Research, 45(6), 3017–3030. [NAR]
---...---
Bio-Synthesis provides a full spectrum of high quality custom oligonucleotide modification services including 5'-triphosphate and back-bone modifications, conjugation to fatty acids, biotinylation by direct solid-phase chemical synthesis or enzyme-assisted approaches to obtain artificially modified oligonucleotides, such as BNA antisense oligonucleotides, mRNAs or siRNAs, containing a natural or modified backbone, as well as base, sugar and internucleotide linkages.
Bio-Synthesis also provides biotinylated mRNA and long circular oligonucleotides".
---...---