Multiplexed error-robust fluorescence in situ hybridization (MERFISH) is a massively parallelized form of the single-molecule fluorescence in situ hybridization (smFISH) method that can image and identify hundreds to thousands of different RNA species simultaneously.
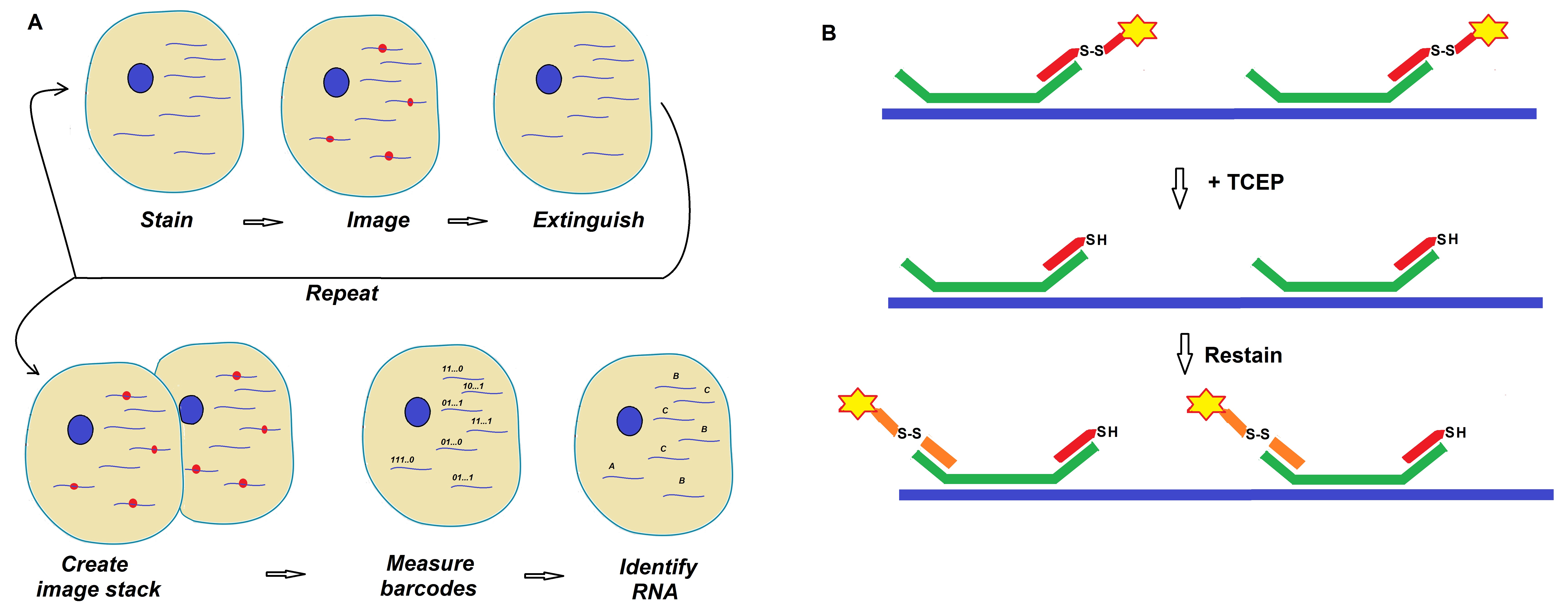
Figure 1: Schematics of a MERFISH readout protocol developed by Moffitt et al. (Moffitt et al. 2016; PNAS).
Imaging based technologies or methods for the study of single-cell transcriptomes have now become very popular. These methods are commonly used as complementary techniques to single-cell RNA sequencing methods. The notion here is that quantitative measurements of the copy number and spatial distribution of fractions of the transcriptome in single cells will revolutionize our understanding of cell and tissue behavior. This knowledge may allow medical researchers to distinguish between healthy and diseased cells and tissues.
Single-molecule fluorescence in situ hybridization (smFISH) is an approach were individual RNAs are labeled using fluorescent probes. If imaging of these species is done in their native context, both copy number, and spatial context can be observed. Moffit et al. developed a technique called multiplexed error-robust fluorescence in situ hybridization (MERFISH) to achieve a higher throughput. This massively parallelized form of smFISH can image and identify hundreds to thousands of different RNA species simultaneously. The use of readout probes allows identification of large numbers of RNAs within a single sample via imaging.
In 2016, Moffit et al. reported that their advancements led to a dramatic increase in throughput using MERFISH. The researchers increased the throughput of MERFISH by two orders of magnitude. Improvements of MERFISH were made using multicolor imaging in combination with the use of chemical cleavage instead of photobleaching. This approach allowed removal of fluorescent signals between rounds of smFISH imaging thereby increasing the imaging field of view.
Using this improved approach the scientists were able to profile 130 RNAs across 40 mm2 of the sample containing approximately 39,000 human cells.
The determination of gene expression is thought to be vital for our understanding of the biology of cells and tissue in different organisms. However, many assays used for the analysis of gene expression destroy the structural context of cells studied.
In recent years, researchers have addressed this issue. The idea is to take a snapshot of switched-on genes in single cells for example in pre-cancerous or cancer cells. Doing this could allow identifying harmful genes at the start of a disease which is crucial for the design of a successful treatment regimen. All human cells carry the same set of genes; however, different genes are switched on in different tissue cells such as muscle versus heart or diseased versus healthy tissue cells. Finding the genes switched on in various diseased cells in their early stages may allow the design of treatments to switch off these cells thereby preventing the development of the disease.
For example, gene expression profiling of tumor cells is now thought to allow the design of less harmful treatments and help to find the best treatment plan.
Recently researchers such as Moffit et al. have combined advances in computational fluorescence microscopy with multiplex probe design allowing simultaneous visualization of gene expression inside single cells with high spatial and temporal resolution.
Readout probe sequences
|
Bit
|
Readout probe
|
Sequence
|
Dye
|
|
1
|
RS0015
|
ATCCTCCTTCAATACATCCC
|
Cy5
|
|
2
|
RS0083
|
ACACTACCACCATTTCCTAT
|
Alexa750
|
|
3
|
RS0095
|
ACTCCACTACTACTCACTCT
|
Alexa750
|
|
4
|
RS0109
|
ACCCTCTAACTTCCATCACA
|
Cy5
|
|
5
|
RS0175
|
ACCACAACCCATTCCTTTCA
|
Cy5
|
|
6
|
RS0237
|
ACCCTTTACAAACACACCCT
|
Alexa750
|
|
7
|
RS0247
|
TTTCTACCACTAATCAACCC
|
Cy5
|
|
8
|
RS0255
|
TCCTATTCTCAACCTAACCT
|
Alexa750
|
|
9
|
RS0307
|
TATCCTTCAATCCCTCCACA
|
Alexa750
|
|
10
|
RS0332
|
ACATTACACCTCATTCTCCC
|
Cy5
|
|
11
|
RS0343
|
TTTACTCCCTACACCTCCAA
|
Cy5
|
|
12
|
RS0384
|
TTCTCCCTCTATCAACTCTA
|
Alexa750
|
|
13
|
RS0406
|
ACCCTTACTACTACATCATC
|
Cy5
|
|
14
|
RS0451
|
TCCTAACAACCAACTACTCC
|
Alexa750
|
|
15
|
RS0468
|
TCTATCATTACCCTCCTCCT
|
Alexa750
|
|
16
|
RS0548
|
TATTCACCTTACAAACCCTC
|
Cy5
|
Note: The dye Is attached to each readout probe via a disulfide bond at the 5′ end of the listed probe sequences.
Reference
Jeffrey R. Moffitt, Junjie Hao, Guiping Wang, Kok Hao Chen, Hazen P. Babcock, and Xiaowei Zhuang; High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. PNAS 2016 : 1612826113v1-201612826.
Wang, G., Moffitt, J.R. & Zhuang, X. Multiplexed imaging of high-density libraries of RNAs with MERFISH and expansion microscopy. Sci Rep 8, 4847 (2018).
-.-