Vaccine development based on messenger RNA (mRNA) is a promising new very useful vaccination approach the production of vaccines against coronaviruses such as SARS-CoV or SARS-Cov-2 (COVID-19) as well as other viruses. mRNAs based vaccines are a promising alternative to conventional vaccines.
The stability of messenger RNAs (mRNAs) influences gene expression in all organisms. In mammals, the abundance of a particular cellular mRNA can fluctuate significantly as the mRNA's half-life changes without any change in transcription. Jess Ross, in a paper published in 1995, discussed the following three questions:
1. Which sequences in mRNAs determine their half-lives?
2. Which enzymes degrade mRNAs?
3. Which factors regulate mRNA stability?
Many mRNAs appear to be degraded by a multistep pathway such that the later step depends on earlier events, for example. The shortening of poly(A) often precedes decay of the mRNA body. For example, the poly(A) tracts of globin mRNAs in young, newly formed developing red blood cells, just entering the circulation, are longer than those in older reticulocytes. Hence, the measured degradation or decay constants (kds) for globin mRNAs can differ in the two reticulocyte populations. As a result, mRNA decay kinetics do not necessarily follow the ideal situations or models. The quantity of full-length polyadenylated and deadenylated mRNA as measured by hybridization assays depends on the mRNA population's shortening rate and age. The cell type can influence the variables and mRNA studied. Sharova et al., in 2009, studied the half-lives of gene expression using whole-genome microarrays in pluripotent and differentiating mouse embrionic stem cells and found that decay rates of mRNAs vary substantially between genes.
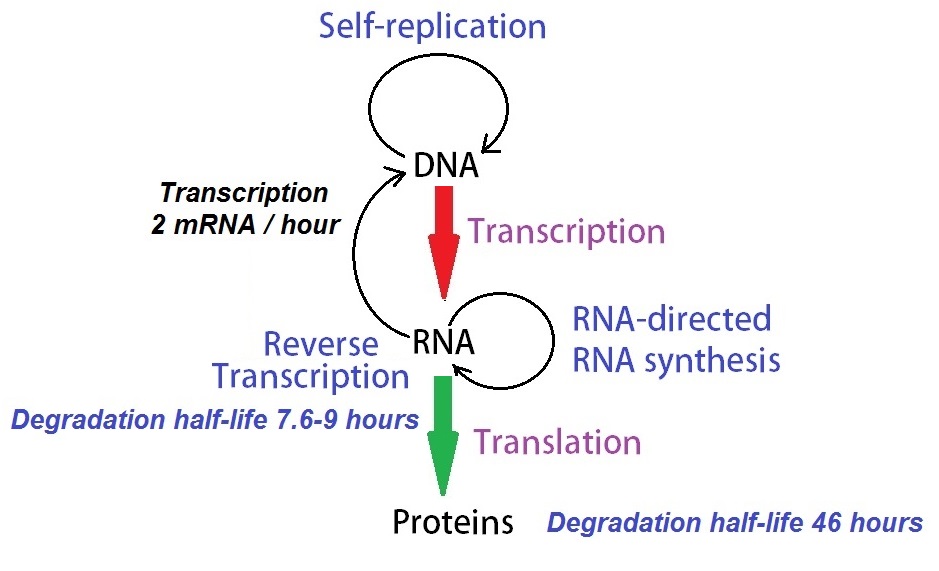
Figure 1: A modified model of Francis Cricks “Central Dogma of Molecular Biology.” Schwanhausser et al. (2011) determined concentrations and degradation rates for 45000 mRNAs and proteins. Their mathematical model using these data describes the cellular dynamics governing protein production. Their analysis showed that ‘transcription is only half the story’, and translational control is as important in determining the final concentrations of proteins.
Factors determining mRNA stability and half-lives
Cis Determinants of mRNA stability
Poly(A): Poly(A) protects mRNAs from rapid degradation.
(i) Deadenylation is the first step in mRNA decay.
(ii) A poly(A)-poly(A)-binding protein (PABP) complex at the mRNA 3’-end protects mRNAs from rapid destruction in vitro.
3’-untranslated regions (UTRs): UTRs influence the half-lives of mRNAs.
(i) Histone mRNA 3’-terminal stem-loop: The 3’-UTRs of histone mRNAs that lack poly(A) affect the processing rates at which the RNA in the nucleus, transported, translated, and degraded. The cell cycle controls these mRNAs. Histone mRNAs (hmRNAs) accumulate to high levels only in S-phase cells. hmRNAs are degraded rapidly at the end of the S phase or when DNA replication is inhibited in S-phase cells. The cis-element at 3'-end on histone mRNAs is responsible for the regulation of histone mRNA degradation.
(ii) AU-rich elements (AUREs): mRNAs containing an AURE and/or an oligo(U) region at a 3’-UTR tend to be unstable. Placing an AURE from the 39-UTR of an unstable mRNA within the 39-UTR of β-globin mRNA, from one encoding granulocyte-macrophage colony-stimulating factor (GM-CSF), the chimeric transcript decays with a half-life of less than 30 min. The 3' untranslated region (UTR) of many messenger RNAs (mRNAs) coding for proto-oncogenes, nuclear transcription factors, and cytokines contain AUREs. ARUEs determine RNA stability in mammalian cells. For example, AUUUA sequences facilitate the degradation of the mRNA body.
(iii) Iron-responsive element (IRE): Transferrin receptor and ferritin encoding mRNAs are regulated post-transcriptionally depending on intracellular iron concentration. The transferrin receptor imports iron into cells. Ferritin is a major iron storage protein in cells. IRE binds an irons-regulatory protein, which depending on its location within the mRNA has different effects. The interaction transferrin receptor and ferritin mRNAs generate iron homeostasis. IREs regulate the half-life of the mRNAs and their translation.
(iv) Long-range stem-loop of insulin-like growth factor II (IGF-II): The insulin-like growth factor II (IGF-II) is necessary for prenatal growth, but its role in adult stem cell physiology is mostly unknown. Ziegler et al., in 2019, demonstrated that IGF-II is critical for multiple adult stem cell niches.
Several sequences in 3’-UTRs influence mRNA stability. 3’-UTRs can also influence mRNA half-lives indirectly, for example, by affecting translation or mRNA localization.
Messenger RNA (mRNA) coding region
The coding region of mRNAs can also determine the half-life of mRNAs.
(i) Mutations in the coding region of mRNAs from c-fos, c-myc, and tubulin can change the half-life. Also, sequences containing protein-binding sites influence the stability of mRNAs and their half-lives.
(ii) Truncated mRNAs lacking most of their 3’-UTRs have half-lives of 1 to 2 hours.
(iii) The introduction of nonsense mutations in the 5’-portion of the coding region destabilizes mRNAs.
5’-Untranslated region, mRNA cap and mRNA localization
5’-UTRs can affect mRNA stability. The introduction of translation-inhibiting stem-loops in the 5’-UTR can change mRNA half-lives several-fold. mRNA half-lives are influences by the length of the UTR, by reciprocal translocations. Longer 5’-UTRs appear to stabilize mRNAs. mRNA without caps are less stable than capped mRNAs, as observed in oocytes and cell-free mRNA decay reactions. Segments affecting the localization of the mRNA can change half-lives as well.
Effector proteins
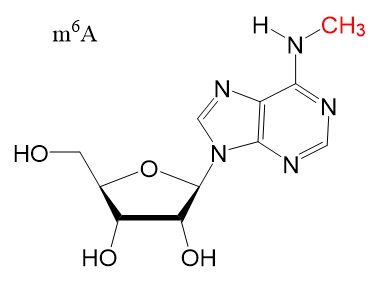
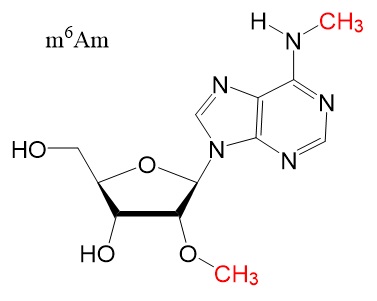
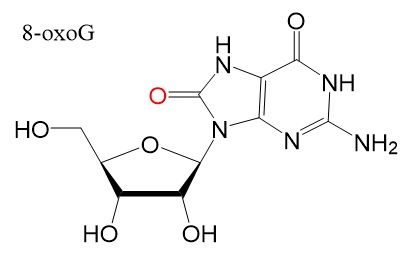
Recent developments in mapping and quantification of RNA modifications demonstrated that modified residues are present in almost all cellular RNA types. Next-generation sequencing (NGS) technologies allowed transcriptome-wide RNA analysis, including modification mapping revealing the presence of significant mRNA modifications in the transcriptome of eukaryotic cells. The identified modifications are N6-methyladenosine (m6A), N6,2'-O-dimethyladenosine (m6Am), 8-oxo-7,8-dihyroguanosine (8-oxoG), pseudouridine (ψ), 5-methylcytidine (m5C), 5-hydroxylmethylcytidine, inosine, and N1-methyladenosine (m1A). Several RNA modifications are now known to regulate mRNA stability. Similar to DNA, RNA can undergo various changes that play a role in cellular and biological processes. The new field studying these modification events in the cell is called "epitranscriptomics."
Effector proteins determine the fate of modified transcripts in a coordinated fashion. These are:
(i) RNA-modifying enzymes that transfer specific chemical groups to a target position on an RNA molecule,
called "Writer Proteins."
(ii) RNA-binding proteins (RBPs) that specifically recognize modified nucleotides, called "Reader Proteins."
(iii) Proteins that remove specific chemical groups form modified nucleotides, called "Eraser Proteins."
Eraser proteins convert modified RNA back to un-modified RNA. Also, endogenous or exogenous chemicals can damage RNA molecules and create modified RNA without involving proteins. Some modifications are reversible, while others are irreversible.
RNA modifications can affect many molecular processes, including transcription, pre-mRNA splicing, RNA export, mRNA translation, and RNA degradation. The regulation of mRNA stability appears to be a crucial step in regulating gene expression. Molecular events involving RNA modifications help to shape the cellular transcriptome and proteome.
Table 1: Chemical structures of RNA modifications affecting mRNA stability.
|
Modification
|
Writers
|
Readers
|
Erasers
|
|
N6-Methyladenosine (m6A)
|
m6A complex composed of methyltransferase like 3 and 14 (METTL3), METTL14, Wilms tumor suppressor gene (WTAP), and KIAA1429
METTL16
|
Stabilization:
IGF2BPs, FMRP, G3BP1, PRRC2A, HuR.
Destabilization:
YTHDF1, YTHGDF2, YTDF3, YTHDC2.
|
ALKBH5
FTO
|
|
N6,2’-O-Dimethyl-adenosine (m6Am)
|
PCIF1
|
Unknown
|
FTO
|
|
8-Oxo-7,8-dihydro-guanosine (8-oxoG)
|
Reactive oxygen species:
Superoxide, hydroxyl radical, hydrogen peroxide.
|
Destabilization: YBX1, AUF1
|
Unknown
|
|
Pseudouridine (ψ)
|
RNA-independent ψ: PUSs
RNA-dependent ψ: Box H/ACA snoRNAs
|
Unknown
|
Unknown
|
|
5-Methylcytidine (m5C)
|
NSUN2
|
Stabilization: YBX1
|
Unknown
|
|
N4-Acetylcytidine (ac4C)
|
NAT10
|
Unknown
|
Unknown
|
Chemical structures
Chemical structure for N6-methyladenosine (m6A), N6,2'-O-dimethyladenosine (m6Am), 8-Oxo-7,8-dihydroguanosine (8-oxoG), pseudouridine (ψ), 5-methylcytidine (m5C), and N4-acetylcytidine (ac4C) are shown below.
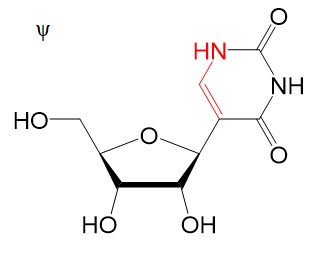
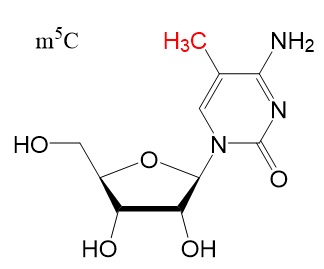
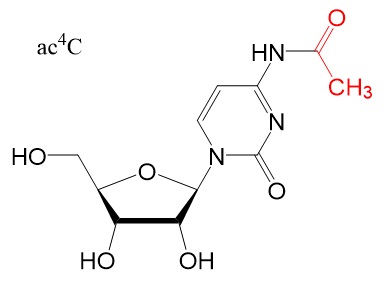
Reference
AU-rich Elements
Boo, S.H., Kim, Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp Mol Med 52, 400–408 (2020). https://doi.org/10.1038/s12276-020-0407-z [Article]
Cobb M (2017) 60 years ago, Francis Crick changed the logic of biology. PLoS Biol 15(9): e2003243.
Crick FHC. On protein synthesis. Symp Soc Exp Biol. 1958;12:138–163. [PubMed]
Li, X., Xiong, X. & Yi, C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 14, 23–31 (2016). [PubMed]
Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5' to 3' and 3' to 5'. Genes Dev. 2008 Jan 1;22(1):50-65. [PMC]
Poly (A) RNA
Ross J.; mRNA stability in mammalian cells. Microbiol Rev. 1995 Sep;59(3):423-50. PMID: 7565413; PMCID: PMC239368. [PMC]
Sharova LV, et al. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. [PMC]
Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473: 337–342. [PubMed]
Schwartz S, Motorin Y. Next-generation sequencing technologies for detection of modified nucleotides in RNAs. RNA Biol. 2017 Sep 2;14(9):1124-1137. [PMC]
Christine Vogel; Translation’s coming of age. Molecular Systems Biology 7; Article number 498. [PMC]
Ziegler AN, Feng Q, Chidambaram S, Testai JM, Kumari E, Rothbard DE, Constancia M, Sandovici I, Cominski T, Pang K, Gao N, Wood TL, Levison SW. Insulin-like Growth Factor II: An Essential Adult Stem Cell Niche Constituent in Brain and Intestine. Stem Cell Reports. 2019 Apr 9;12(4):816-830. [PMC]
---...---
Bio-Synthesis provides a full spectrum of high quality custom oligonucleotide modification services by direct solid-phase chemical synthesis or enzyme-assisted approaches to obtain artificially modified oligonucleotides containing backbone, base, sugar and internucleotide linkages including messenger RNA.
Bio-Synthesis specialize in complex oligonucleotide modifications using phosphodiester backbone, purine and pyrimidine heterocyclic bases, and sugar modified nucleotides such as our patented 3rd generation Bridged Nucleic Acids.