Poly-A refers to the poly-A tail at the 3'-end of oligonucleotides consisting of a repetitive sequence of adenine nucleotides. The 3'-end of nearly all eukaryotic mRNAs include a string of 50 to 250 adenylate residues, called poly-A tail. Poly-A is added posttranscriptional to the 3'-end of a eukaryotic mRNA molecule. Poly-A polymerase generates the poly-A tail from adenosine triphosphate (ATP). The addition of a poly-A tail to mRNAs is called "polyadenylation." Most mRNAs contain a poly-A tail. However, structural RNAs do not. In eukaryotes, the 5' cap and poly(A) tail of mRNAs together enhance translation through a process that requires the interaction of the cap-associated eukaryotic initiation factor, eIF-4G, and the poly(A)-binding protein, PABP.
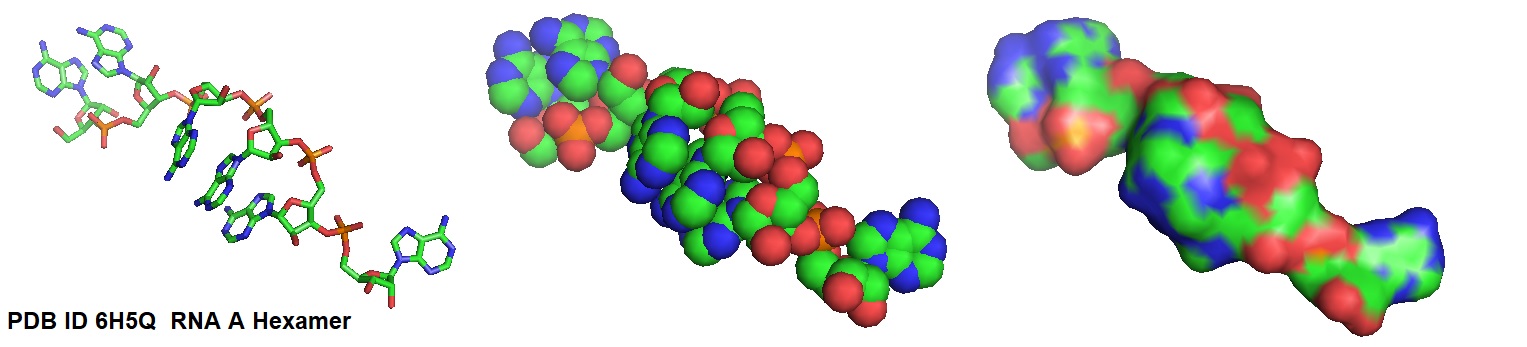
Figure 1: Structural models of a poly A hexamer. The structure of this RNA poly A hexamer was solved via cryo-EM of a nucleocapsid RNA complex from measles virus nucleocapsid-like particles on specific sequences of RNA hexamers (poly-Adenine and viral genomic 5') in vitro [Desfosses et al., 2019; PDB ID 6H5Q].
The presence of the poly-A tail allows easy separation of mRNAs from more prevalent ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) present in cell extracts. Purification columns containing a matrix or beads such as magnetic beads linked to short strings of thymidylate or oligo-dTs allow trapping and purification of RNA molecules containing a poly-A tail. When passing a cell extract though an oligo-dT column, the poly-A tails base-pair with the oligo-dTs and bind the mRNAs to the column. The RNAs that do not attach to the column can be washed away. Bound mRNAs are recovered by elution with a low-salt buffer. This method has proved essential for the construction of cDNA libraries.
Oligonucleotides modified with bridged nucleic acids (BNAs), a class of bicyclic RNA analogs, have a high affinity for their complementary DNA and RNA targets. BNA-oligo-T-capture probes allow isolation and purification of poly-A RNA from guanidine thiocyanate-lysed cell extracts. The enzyme reverse transcriptase, found in retroviruses, can synthesize a complementary strand to each mRNA molecule. The enzyme can only add nucleotides to the 3’-end of a preexisting primer base-paired to a template. Added free oligo-dT can serve this function by hybridizing to the 3’-poly-A tail of each mRNA template.
Processing in higher eukaryotes starts from the initial RNA product synthesized by RNA polymerase II. The first RNA products called “primary transcripts” undergo several processing steps before the production of functional mRNAs. RNA polymerase initiates transcription at the first nucleotide of the first exon of a gene. Capping of the 5’-end of the nascent RNA with 7-methyl-guanylate occurs just after transcription begins. Processing occurs in the nucleus resulting in a functional, mature mRNA, transported to the cytoplasm. The 3’-end of a functional mRNA is generated by endonucleolytic cleavage at a specific sequence, the poly-A site, located at the 3’-end of the final exon. After cleavage, a stretch of 100 to 250 adenine residues is added to the 3’-hydroxyl group. RNA splicing removes introns before the transportation of completed mRNA to the cytoplasm.
In animal cells, all mRNAs, except histone mRNAs, have a 3’-poly-A tail. The poly-A-binding protein associated with mRNA poly-A tails appears to interact with actin microfilaments of the cytoskeleton. poly(A) tails, in eukaryotes, usually act as stabilizers of intact mRNAs, whereas in E. coli they serve to accelerate the destruction of fragments.
Degradation of mRNAs modulates gene expression, and quality controls mRNA biogenesis. The 3’- poly-A tails of almost all eukaryotic mRNAs have two significant functions.
(i) Control of mRNA decay. The degradation of the poly(A) tail by a 3′ exonuclease called deadenylation is the first step
in mRNA decay.
(ii) Completion of deadenylation triggers the second step: cap hydrolysis or further 3′ to 5′ degradation.
The nuclear poly(A)-binding protein (PABPN1) controls the length of the poly-A tail. PABPN1 regulates the interaction between poly-A polymerase and the cleavage and polyadenylation specificity factor.
Reference
Bernstein P, Ross J. Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem Sci. 1989;14(9):373-377. [TIBS]
Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA. 2011 Mar-Apr;2(2):167-83. [PMC]
Desfosses A, Milles S, Jensen MR, Guseva S, Colletier JP, Maurin D, Schoehn G, Gutsche I, Ruigrok RWH, Blackledge M.; Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication. Proc Natl Acad Sci U S A. 2019 Mar 5;116(10):4256-4264. [PNAS]
Marc Dreyfus and Philippe Régnier; The Poly(A) Tail of mRNAs: Minireview Bodyguard in Eukaryotes, Scavenger in Bacteria. Cell, Vol. 111, 611–613, November 27, 2002. [Cell]
Bio-Synthesis Inc. is pleased to offer a large variety of DNA/RNA oligonucleotides and peptides, modified or unmodified including poly A and poly T oligonucleotides, various conjugates, as well as stapled peptides and regular peptides, for a number of research applications, including COVID 19 testing, analysis and vaccine development!
---...---