The development of the 2-(2-nitrophenyl) propoxy-carbonyl (NPPOC) group permitted the manufacture of microarrays of long oligonucleotides. The NPPOC photolabile protecting group essentially allows quantitative deprotection with a high photolysis quantum yield during the manufacture of microarrays of long oligonucleotides.
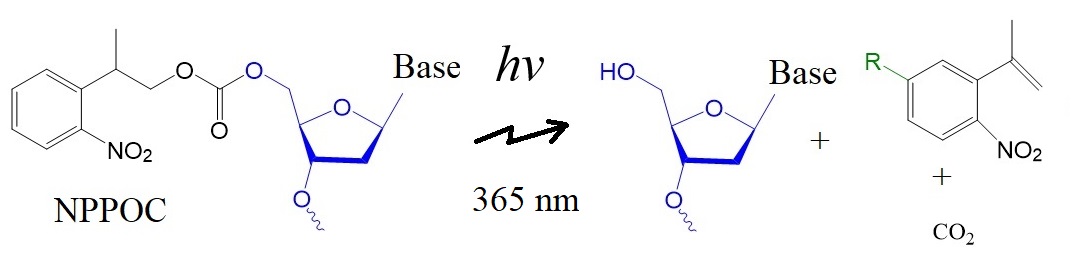
Figure 1: Structure of the photolabile NPPOC protecting group and its photocleavage products.
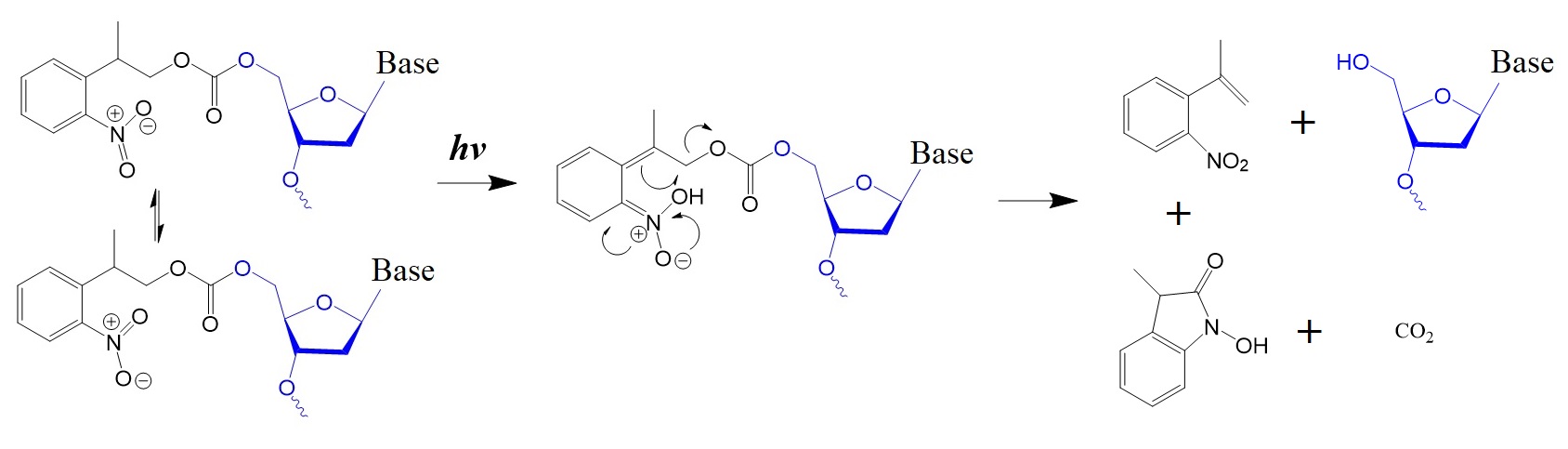
Figure 2: Proposed photo-deprotection mechanism of NPPOC.
According to Hasan et al. (1997), Buehler et al. (2004), and Chen et al. (2022), the photocyclization of NPPOC to the N-hydroxy indole ketone involves the initial formation of nitrostyrene products that could cyclize with the cleavage of the Cβ-O bond because several of the side products are inactive. This reaction is advantageous in the synthesis of oligonucleotides and cyclic peptides.
NPPOC or 2-(2-nitrophenyl)-propoxy-carbonyl oligonucleotides are photolabile oligonucleotides useful in molecular biology and genetics research. NPPOC oligonucleotides contain the photolabile nitrophenyl)-propoxy-carbonyl group at the 2'-position of the ribose sugar. BzNPPOC oligonucleotides have the more labile 2-(2-nitrophenyl)-propoxy-carbonyl group attached at the 2'-position. NPPOC oligonucleotides are chemically modified nucleic acids that offer selective protection, reversible deprotection, and stability. Their versatility makes them valuable tools for various molecular biology applications, including oligonucleotide synthesis, library construction, and antisense technologies.
The availability of commercial NPPOC phosphoramidites now enables automated custom solid phase synthesis of NPPOC modified oligonucleotides.
Light as an external trigger signal allows for controlled direct chemical methods with high spatial and temporal accuracy. Photolabile protecting groups can be removed with light allowing for a high degree of chemo-selectivity. The nitrobenzyl-modified photolabile protecting groups are the most used.
Photolabile groups extend available orthogonal protecting strategies to enable technics such as photopolymerization, cross-linking, and functionalization in polymer chemistry for 3D patterning and fabrication, as well as creating biologically inactivated (caged) molecules. After the introduction into cells, a beam of light can activate caged molecules. Caged adenosine triphosphate (ATP) photolysis is an excellent example of a controlled photo-releasable reaction. Caged ATP [NPE-caged ATP; P3-(1-(2-nitrophenyl)-ethyl-adenosine 5’-triphosphate] is a nucleotide analog containing a blocking or protecting group at the terminal phosphate group, the γ-phosphate. The presence of the blocking group renders the molecule biologically inactive. Flash photolysis of the blocking or caging group with UV light illumination at around 360 nm rapidly releases the caging group, releasing the free nucleotide locally.
Photolabile groups enable high-resolution spatial control of reactions when optical imaging systems deliver the light. Spatial control is beneficial for the combinatorial synthesis of biopolymer microarrays. This approach can produce microarrays with >106 unique sequences per square centimeter.
|
Hasan et al., in 1997, reported the development of photosensitive protecting groups for nucleoside 5’-hydroxyl group containing 2-(2-nitrophenyl)-ethoxy-carbonyl groups. The researchers reported the synthesis of a series of thymidine derivatives with 5’-photolabile protecting groups. During this study, the research group identified the 2-(2-nitrophenyl)-propoxy-carbonyl (NPPOC) group as a particularly promising candidate.
Giegrich et al., in 1998, showed that the NPPOC group can be readily removed from 5’-O-protected thymidine derivatives by irradiation at 365 nm.
In 1997, Dell’Aquilla et al. reported the use of the o-nitro-phenyl-1,3-propanediol group for the synthesis of a solid-phase support for automated oligonucleotide synthesis.
The research group covalently linked o-nitrophenyl-1,3-propanediol to long-chain alkyl amine-controlled pore glass (LCAA-CPG) beads through a stable phosphoramidate diester link. This solid support allowed the automated synthesis of 3′-phosphate unmodified and base-sensitive oligonucleotides. Photolytic cleavage released the final oligonucleotides from the solid support with high efficiency.
|
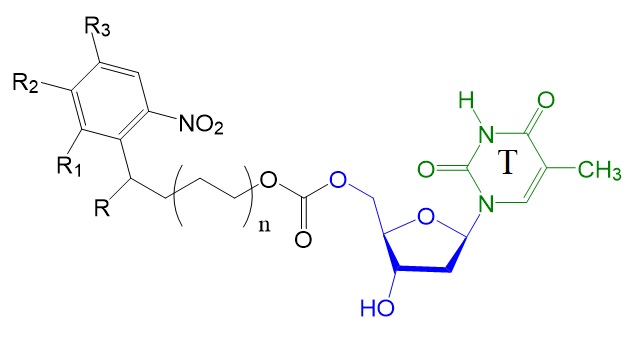
Figure 3: Photolabile thymidines investigated by Hasan et al. in 1997.
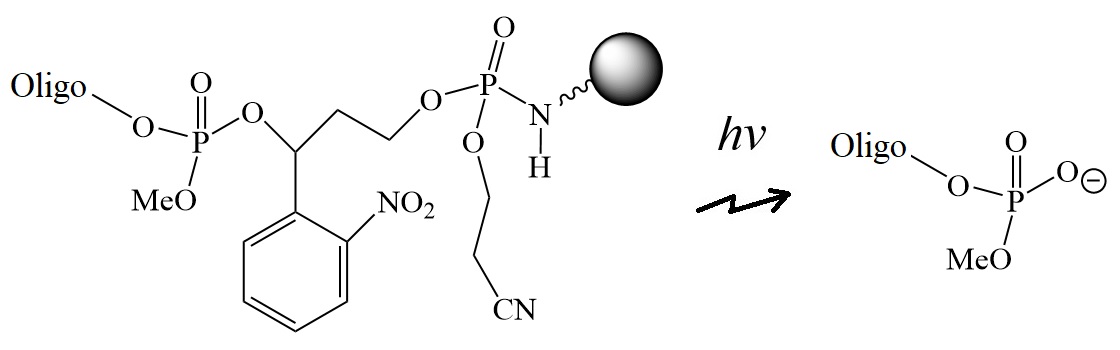
Figure 4: o-Nitrophenyl-1,3-propandial long chain alkyl amine controlled pore glass.
|
The NPPOC group is now widely used for synthesizing genomic DNA microarrays, the synthesis of aptamers, gene assembly, RNAs, and peptide microarrays, in carbohydrate chemistry, as cleavable linkers, and for caging. The photolysis quantum yield of NPPOC is relatively high (0.41 in MeOH). Still, its low absorptivity (ε365nm/MeOH ≈ 230 M−1 cm−1) has led to both the search for derivatives with higher absorptivity and the development of photosensitization techniques based on intra- and intermolecular energy transfer from a triplet sensitizer. A triplet sensitizer absorbs a low-energy photon and generates a singlet exciton that is rapidly converted into a triplet exciton via intersystem crossing to transfer the triplet exciton to an emitter material with low-lying triplet and high-lying singlet state energies.
Introducing the 2-(2-nitrophenyl)-propoxy-carbonyl (NPPOC) or benzoyl-2-(2-nitrophenyl)-propoxy-carbonyl (Bz-NPPOC) group into oligonucleotides offers several advantages for various applications. The NPOC modification allows selective protection of the 2'-hydroxyl group, which is crucial for controlling the reactivity and stability of oligonucleotides. Masking the 2'-hydroxyl group with the NPPOC moiety prevents unwanted chemical reactions or enzymatic degradation at that position.
The synthesis of NPPOC oligonucleotides involves coupling a 2-(2-nitrophenyl)-propoxy-carbonyl phosphoramidite building block to the growing oligonucleotide chain during solid-phase synthesis. This step introduces the NPPOC modification at the desired position, and subsequent deprotection and purification steps yield the final NPPOC oligonucleotide product.
The NPPOC modification can be selectively removed under mild conditions, allowing the recovery of the unmodified 2'-hydroxyl group for further functionalization or conjugation reactions. This flexibility makes NPPOC oligonucleotides highly versatile in a range of applications. For example, deprotection of NPPOC enables enzymatic ligation, which is particularly useful in assembling longer oligonucleotides or synthesizing nucleic acid constructs allowing for various solid-phase synthesis strategies. The reversible nature of the NPPOC modification enables iterative coupling and deprotection steps, useful for the synthesis of complex oligonucleotide libraries or combinatorial structures.
The NPPOC modification enhances the stability and resistance of oligonucleotides against nuclease degradation, ensuring their prolonged activity in biological systems.
Examples of applications and studies
Beier and Hoheisel, in 2000, reported the photolithographic synthesis of DNA chips utilizing 5’-[2-(2-nitrophenyl)-propyloxycarbonyl]-2’-deoxynucleoside (NPPOC) phosphoramidites. The efficient quantitative photo-deprotection step influenced the quality of the resulting DNA chips. This approach resulted in an increase of synthesis yields by more than 10-fold for 20mer oligonucleotides. The researchers pointed out that DNA chips enable hybridization applications.
Pirrung et al., 2001, developed pyrimidine building blocks for 5’-3’ DNA synthesis utilizing the NPPOC protecting groups to allow for automated photochemical DNA synthesis in a synthesizer.
Blair et al., in 2006, reported the synthesis of DNA oligonucleotide strands in capillaries utilizing photolabile NPPOC-chemistry and ultraviolet-light emitting diodes (UV-LEDs). The researchers synthesized multiple oligonucleotides in single capillaries and characterized the final products via hybridization, sequencing, and gene synthesis. The capillary-based DNA synthesis produced functional oligonucleotides with up to 44% perfect sequences. According to the researchers, the capillary-based synthesis system offers a novel, scalable approach for synthesizing high-quality oligonucleotides useful for biological applications but may need improvements to allow the synthesis of longer oligonucleotides.
Wöll et al., in 2007, studied various covalently linked thioxanthone (TX)−linker−2-(2-nitro-phenyl)-propoxy-carbonyl (NPPOC)−substrate conjugates where the TX chromophore functioned as an intramolecular sensitizer to the NPPOC moiety. Quantitative stationary fluorescence spectroscopy allowed the researchers to determine the rate of electronic energy transfer between TX and NPPOC. The study observed a dual mechanism of triplet−triplet energy transfer encompassing a slower mechanism involving the T1(ππ*) state of TX with linker-length-dependent time constants longer than 20 ns and a fast mechanism with linker-length-dependent time constants shorter than three ns. The latter mechanism involved the energy transfer from the T2(nπ*) state, which is in fast equilibrium with the fluorescent S1(ππ*) state. The spectroscopic results revealed the presence of one united chromophore, which shows the typical NPPOC cleavage reaction triggered by intramolecular hydrogen atom transfer to the nitro group.
Agbavwe et al., in 2011, studied efficiencies, errors, and yields for the light-directed maskless synthesis (MAS) of DNA microarrays. During MAS array synthesis, the phosphoramidite chemistry achieved coupling efficiencies comparable to solid-phase oligonucleotide synthesis of ~99%. The reported increased coupling efficiency allowed the synthesis of 60mers.
Light-directed synthesis of microarrays is a photolithographic technology borrowed from the semiconductor industry combined with combinatorial chemistry of phosphoramidites utilizing a photolabile 5’-hydroxyl protecting group. In 1991, Fodor et al. showed that solid-phase chemistry, photolabile protecting groups, and photolithography could be combined to achieve light-directed, spatially addressable parallel chemical synthesis to produce a highly diverse set of chemical products.
The MAS method is similar to conventional solid-phase synthesis of oligonucleotides, however, the synthesis of microarrays is more complex. The unique synthesis kinetics on the glass substrate requires careful tuning of parameters and modifications to the synthesis cycle to achieve optimal deprotection and phosphoramidite coupling.
Franssen et al., in 2013, demonstrated that light-activatable aptamers enable the production of microarrays. Franssen et al. utilized in situ synthesis to synthesize oligonucleotide microarrays and suggested increasing the spacer length and maximizing oligonucleotide sequence fidelity can significantly improve aptamer microarray detection.
According to the study, aptamer microarrays are less sensitive than hybridization microarrays to molecular crowding. However, the functionalization chemistry of the glass substrate affected the aptamer binding signal, either by modifying the oligonucleotide surface density or via electrostatic or hydrophobic interactions with the aptamers or target protein. According to Franssen et al., aptamer microarrays are a promising high-throughput method for ultrasensitive detection of multiple analytes.
More recently, Kretchy et al., in 2015, introduced two more o-nitrobenzyl derivatives of the NPPOC group with improved photo-deprotection efficiencies. The structures of oligonucleotides modified with these photo-cleavable groups are illustrated in figure 4.
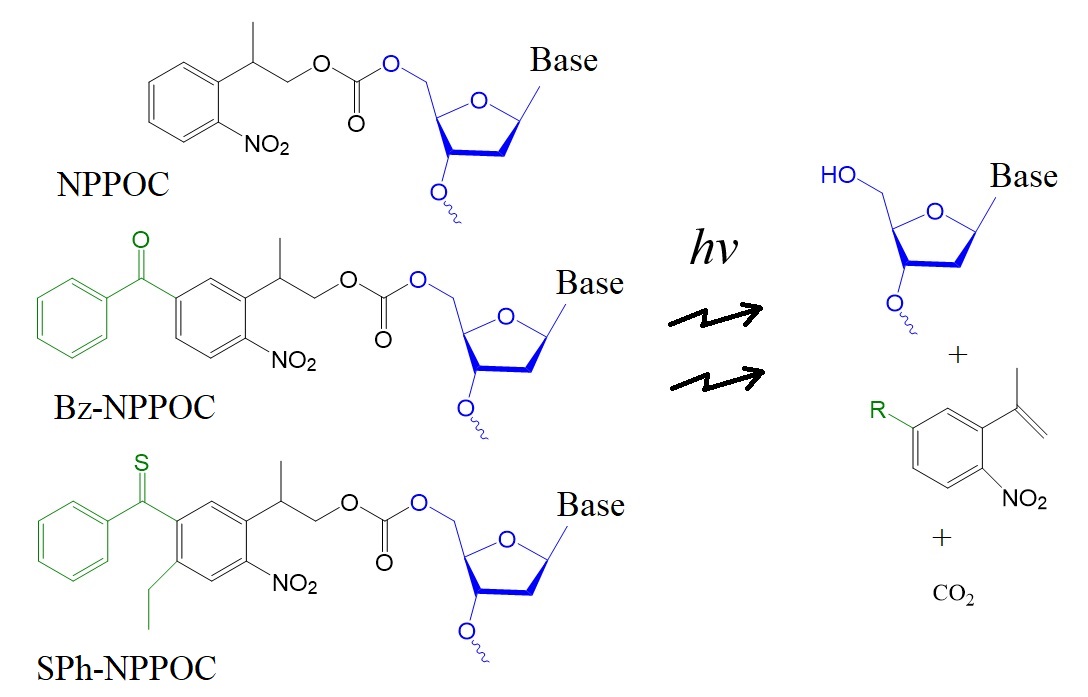
Figure 5: Structures and photocleavage products of DNA oligonucleotides modified with NPPOC, Bz-NPPOC, and SPh-NPPOC 5'-OH protecting groups on the 5’-terminal ends.
Hoelz et al., in 2018, optimized a photolithographic in situ maskless array synthesizer system for the fabrication of 5′→3′ oligomers. Regular in situ synthesized DNA arrays is done in the traditional 3′→5′ direction. However, recently emerged new applications, such as spatial transcriptomics and enzymatic synthesis of RNA, require a reverse, 5′→3ʹ oriented array synthesis or combined 5′→3′ and 3′→5′ synthesis.
Reference
Agbavwe C, Kim C, Hong D, Heinrich K, Wang T, Somoza MM. Efficiency, error and yield in light-directed maskless synthesis of DNA microarrays. J Nanobiotechnology. 2011 Dec 8;9:57. doi: 10.1186/1477-3155-9-57. [PMC]
Beier M, Hoheisel JD. Production by quantitative photolithographic synthesis of individually quality checked DNA microarrays. Nucleic Acids Res. 2000 Feb 15;28(4):E11. [PMC]
Blair S, Richmond K, Rodesch M, Bassetti M, Cerrina F. A scalable method for multiplex LED-controlled synthesis of DNA in capillaries. Nucleic Acids Res. 2006;34(16):e110. doi: 10.1093/nar/gkl641. Epub 2006 Sep 8. Erratum in: Nucleic Acids Res. 2007;35(2):703. [PMC]
Bühler S., Lagoja I., Giegrich H., Stengele K.P., Pfleiderer W. New Types of Very Efficient Photolabile Protecting Groups Based upon the [2-(2-Nitrophenyl)Propoxy]Carbonyl (NPPOC) Moiety. Helv. Chim. Acta. 2004; 87:620–659. [Helvetica]
Chen T, Wang G, Tang L, Yang H, Xu J, Wen X, Sun Y, Liu S, Peng T, Zhang S, Wang L. Synthesis of Cyclic Peptides in SPPS with Npb-OH Photolabile Protecting Group. Molecules. 2022 Mar 29;27(7):2231. doi: 10.3390/molecules27072231. [PMC]
Dell'Aquila, Ch., Imbach, J.-L., Rayner, B.; Photolabile linker for the solid-phase synthesis of base-sensitive oligonucleotides. Tetrahedron Letters, 38, 30, 1997, 5289-5292. [Tetrahedron Letters]
Franssen-van Hal NL, van der Putte P, Hellmuth K, Matysiak S, Kretschy N, Somoza MM. Optimized light-directed synthesis of aptamer microarrays. Anal Chem. 2013 Jun 18;85(12):5950-7. [PMC]
Fodor S, Read J, Pirrung M, Stryer L, Lu A, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991; 251:767–773. [PubMed]
Giegrich H., Eisele-Bühler,S., Hermann,C., Kwasyuk,E., Charubala,R. and Pfleiderer,W. (1998) New Photolabile Protecting Groups in Nucleoside and Nucleotide Chemistry—Synthesis, Cleavage Mechanisms and Applications. Nucleosides, Nucleotides & Nucleic Acids. Nucl. Nucl., 17, 1987–1996. [NN&N]
Hasan A, Stengele KP, Giegrich H, Cornwell P, Isham KR, Sachleben RA, Pfleiderer W, Foote RS. Photolabile protecting groups for nucleosides: Synthesis and photodeprotection rates. (1997) Tetrahedron. 53: 4247-4264. [Tetrahedron]
Hölz, K., Hoi, J., Schaudy, E. et al. High-Efficiency Reverse (5′→3′) Synthesis of Complex DNA Microarrays. Sci Rep 8, 15099 (2018). [Nature]
Jemt, A. et al. An automated approach to prepare tissue-derived spatially barcoded RNA-sequencing libraries. Sci Rep 6, 37137, (2016). [Scientific Reports] Derived spatially barcoded RNA-sequencing libraries.
Kretschy N, Holik AK, Somoza V, Stengele KP, Somoza MM. Next-Generation o-Nitrobenzyl Photolabile Groups for Light-Directed Chemistry and Microarray Synthesis. Angew Chem Int Ed Engl. 2015 Jul 13;54(29):8555-9. [PMC]
Pirrung MC, Wang L, Montague-Smith MP. 3'-nitrophenylpropyloxycarbonyl (NPPOC) protecting groups for high-fidelity automated 5' --> 3' photochemical DNA synthesis. Org Lett. 2001 Apr 19;3(8):1105-8. [PubMed] -> 5'->3'-Synthesis.
Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg Å, Pontén F, Costea PI, Sahlén P, Mulder J, Bergmann O, Lundeberg J, Frisén J. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016 Jul 1;353(6294):78-82. [PubMed] -> Spatial transcriptomics.
Vickovic, S. et al. Massive and parallel expression profiling using microarrayed single-cell sequencing. Nat Commun 7, 13182, (2016). [Nature Communications] -> Microarrayed single-cell sequencing.
Wöll D, Laimgruber S, Galetskaya M, Smirnova J, Pfleiderer W, Heinz B, Gilch P, Steiner UE. On the mechanism of intramolecular sensitization of photocleavage of the 2-(2-nitrophenyl)propoxycarbonyl (NPPOC) protecting group. J Am Chem Soc. 2007 Oct 10;129(40):12148-58. [JACS]
Wu, C.-H., Holden, M. T. & Smith, L. M. Enzymatic Fabrication of High-Density RNA Arrays. Angew Chem, Int Ed 53, 13514–13517. (2014). [PMC] -> Enzymatic Fabrication.
--...--