The role of mRNA in Gene expression
Gene expression refers to the conversion of genetic information from genes via messenger RNA (mRNA) to proteins. The entire set of genes in an organism is called the genotype. This set of genes includes alleles that when expressed determine the trait or phenotype of an organism. According to the central dogma of molecular biology this conversion occurs via transcription to generate mRNA followed by translation to produce the gene product, usually a protein. The result of the total sum of expressed genes in an organism manifests the phenotype of the individual organism. However, gene expression of genes that do not code for proteins only involves transcription. This is the case for ribosomal RNA (rRNA) and transfer RNA (tRNA), as well as other RNA molecules involved in regulatory pathways. Messenger RNA is the key molecule to enable gene expression for the production of proteins. A structural model of a mature messenger RNA is illustrated in figure 1.

Figure 1: Mature messenger RNA.
Messenger RNA (mRNA) is the molecule that links genes to proteins. Efficient and smooth interactions of the molecules of life allow us humans to function well. Major parts of a human body, as well as any mammalian body, visible to human eyes, are made up of proteins. However, for a human cell to function, more than proteins are needed. The coordinated interplay of nano-machines consisting of proteins, DNA, RNA, metal ions and other metabolic molecules are needed for a cell to function. On the molecular level, most of a human’s daily functions are executed by proteins. For the production of proteins in cells, messenger RNA is needed. According to the “central dogma of molecular biology” double-stranded DNA (dsDNA) located in the cell nucleus is transcript into RNA complementary to the DNA template it originates from and is further translated into protein sequences made up of amino acids to produce functional proteins. This process is called gene expression. As humans and their cells age body protein synthesis change with changes occurring in their metabolism. The rate of protein synthesis per kg total body weight is known to decline with growth and development within a species.
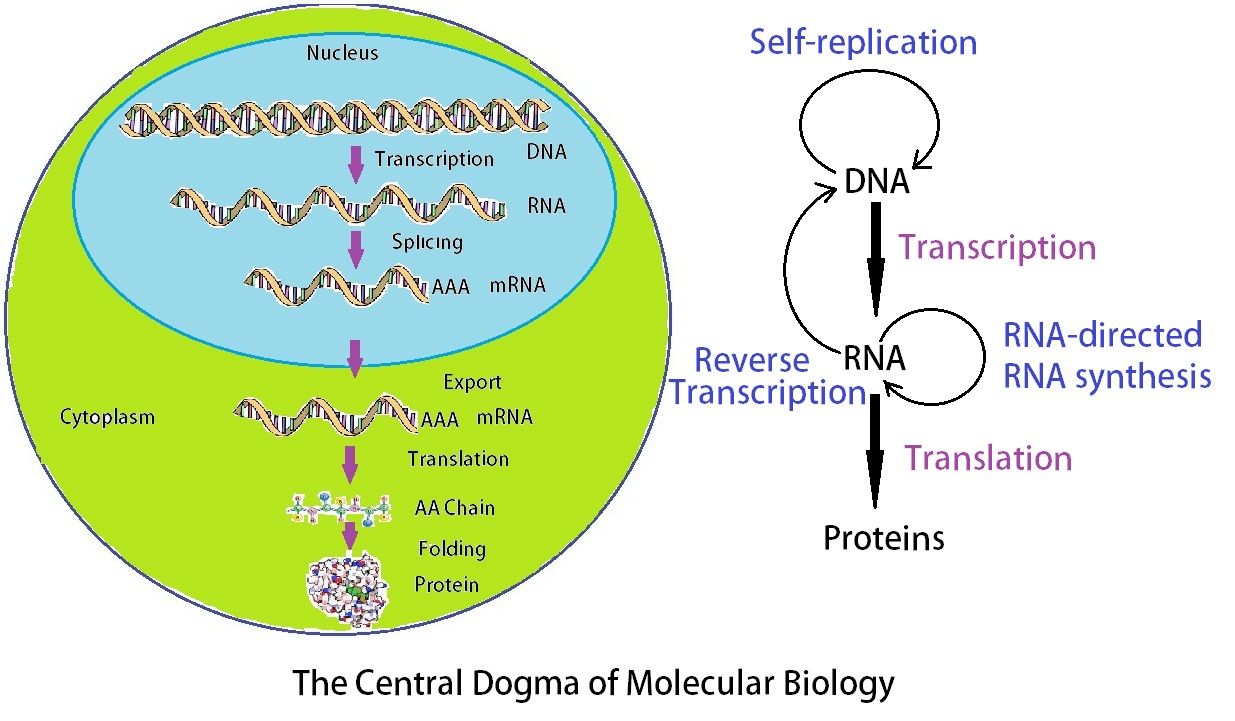
Figure 2: The central dogma of molecular biology as stated by Francis Crick explains the flow of genetic information within a biological system. Simplistically it states: “DNA makes RNA and RNA makes protein.” However, it has become apparent in recent years that this simplistic view is only part of the story of gene expression.
Gene expression is known to occur via a two-stage process – transcription and translation.
Transcription
Transcription generates a single-stranded RNA (ssRNA) identical to one sequence stretch in the strands of duplex DNA. However, different types of RNA are generated by transcription. Protein synthesis in cells involves three principal classes: messenger RNA (mRNA), transfer RNA (tRNA), and ribosomal RNA (rRNA).
Translation
Translation converts the nucleotide sequence of RNA into the sequence of amino acids needed for the synthesis of a protein. An mRNA is translated into a protein sequence. tRNA and rRNA are other needed parts for the protein synthesis apparatus. However, the entire length of the mRNA is not translated. Each mRNA contains, at least, one coding region that is related to a protein sequence of the genetic code. Each nucleotide triplet called codon of the coding region represents one amino acid. Only one strand of a DNA duplex is transcribed into a messenger RNA molecule. The DNA strand that directs the synthesis of mRNA is called the template or antisense strand. mRNA synthesis occurs via complementary base pairing. The term “antisense” is used to describe a sequence of DNA or RNA that is complementary to mRNA. The DNA strand that contains the same sequence as the mRNA is called the coding strand or sense strand. However, it possesses T instead of U.
The genetic code is read on the mRNA which is usually described in terms of the four bases present in RNA. These are U, C, A, and G. Messenger RNA is translated by ribosomes. The ribosome catalyzes the translation of mRNA into polypeptide chains. Historically ribosomes are described by their sedimentation constant S. This constant S (for Svedberg) indicates the rate of sedimentation for the ribosome investigated. A greater rate of sedimentation indicates a larger mass. The ribosome is a ribonucleoprotein particle consisting of two subunits that work together as part of the complete ribosome.
Advances made in next-generation DNA sequencing and proteomics now provide the ability to survey mRNA and protein abundances. These proteome-wide surveys now enable determination of the extent to which different aspects of gene expression help regulate cellular protein abundances. Therefore, it has now become clear that substantial regulatory processes occur after mRNA is made. Recent studies indicate that post-translational, translational and protein degradation based regulation events control the steady-state of protein abundances. The steady-state of protein abundance refers to the condition of a cell or biological system in which the abundance of proteins stays constant or changes only negligibly over time.
The biological concentrations of proteins in cells are regulated by interactions of transcription, translation, mRNA and protein degradation. In 2011 Schwanhausser et al. measured concentrations of mRNAs and proteins and the corresponding degradation rates for >5,000 genes in mouse cells. Using a mathematical model, the researchers described the dynamics that govern mammalian protein production (see figure 3 below).
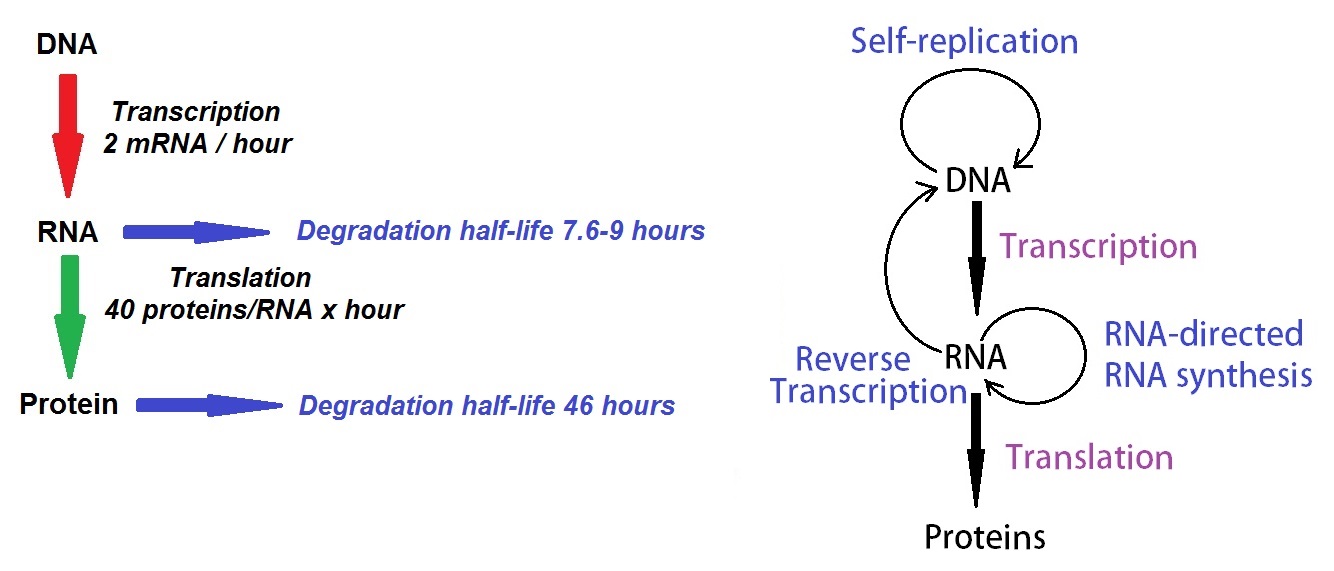
Figure 3: Revised description of the central dogma of molecular biology as first described by Francis Crick.
Reference
Lewin, Benjamin; Genes VII, 2000, Oxford University Press ISBN 0-19-979276-X (Hbk). pp 119 – 190.
VERNON R. YOUNG, WILLIAM P. STEFFEE, PAUL B. PENCHARZ, JOERG C. WINTERER & NEVIN S.SCRIMSHAW; Total human body protein synthesis in relation to protein requirements at various ages. Nature 253, 192 - 194 (17 January 1975); doi:10.1038/253192a0.
Vogel, Christine, and Edward M. Marcotte. “Insights into the Regulation of Protein Abundance from Proteomic and Transcriptomic Analyses.” Nature reviews. Genetics 13.4 (2012): 227–232. PMC. Web. 28 Jan. 2016.
Schwanhausser, Bjorn, Busse, Dorothea, Li, Na, Dittmar, Gunnar, Schuchhardt, Johannes, Wolf, Jana, Chen, Wei, Selbach, Matthias; Global quantification of mammalian gene expression control. Nature 2011/05/19/print 473, 7347, 337-342.