Threofuranosyl nucleotides are the building blocks of α-l-threose nucleic acids (TNAs). The TNA moiety has one less atom in its backbone than the DNA backbone. The B-family polymerase Kod-RI, a laboratory-evolved polymerase derived from the archaeal hyperthermophilic species Thermococcus kodakarensis (Kod), allows chemical synthesis of TNAs.
| 
|
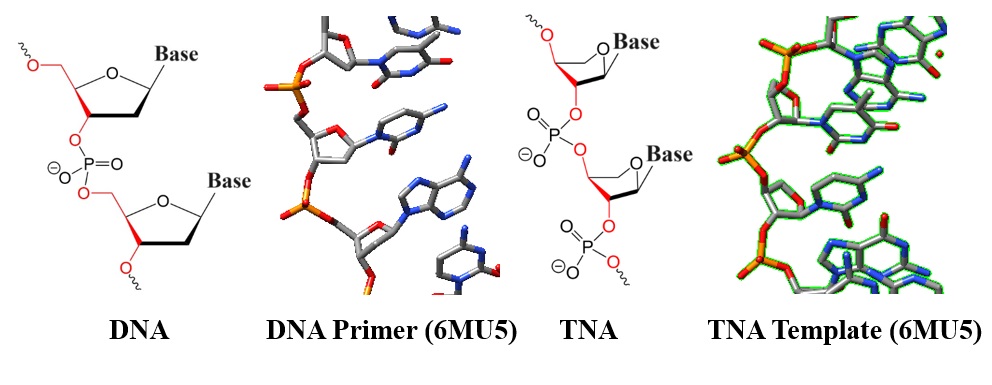
| Figure 1: Chemical structures and models of DNA and TNA. |
Components of TNA are
Threofuranose: A threofuranose moiety is part of the sugar backbone of threofuranosyl nucleotides. Unlike the ribose sugar in RNA and deoxyribose in DNA, threofuranose has four carbon atoms arranged in a furan ring, resulting in a slightly different shape. TNA oligomers contain α-L-threofuranosyl nucleotide repeats connected by 3′-,2′-phosphodiester linkages.
Nucleobase: Like RNA and DNA, threofuranosyl nucleotides also have nucleobases attached to the sugar. These can include adenine (A), guanine (G), cytosine (C), and thymine (T), uracil (U) or modified bases.
Phosphate group: The phosphate group connects the threofuranosyl nucleotide units to form TNA strands.
Unique Features of TNA
Backbone arrangement: TNA strands have a different backbone linkage than RNA and DNA (see chemical structures in Figure 1). The phosphate group connects the 2'-carbon of one threofuranose to the 3'-carbon of the next instead of the usual 3'- to 5'-linkage in RNA and DNA.
Hybridization: Despite the different backbone, TNA can still form double helices by pairing with itself, RNA, and DNA. These characteristics make it a potential evolutionary alternative to RNA that might have played a role in the early stages of life.
Applications of TNA
As a research tool: TNA's unique properties make it a valuable tool for studying the evolution of nucleic acids and understanding how information storage and replication work.
As potential biomedicals: Scientists are exploring TNA's potential for designing new drugs and gene therapy tools, for example, TNA aptamers or XNAs. Its stability and ability to resist enzymes make it an attractive candidate for these applications, particularly for developing new aptamer types.
TNA libraries: Engineered polymerases allow the synthesis of TNA libraries, enabling in vitro selection of sequences that bind to ligands or have catalytic activity.
TNA-based enzymes for gene silencing: Engineered TNA enzymes enable selective gene silencing. Wang et al., in 2022, report the in vitro selection of RNA-cleaving TNA enzymes. The research group biochemically characterized one of the selected enzymes called Tz1 and showed that Tz1 exhibited a strong sequence selectivity toward its RNA substrate. The researchers derived the substrate from a mutant epidermal growth factor receptor (EGFR) mRNA involved in drug resistance. Also, Wang et al. showed that Tz1 selectively silenced mutant EGFR gene expression in a non-small-cell lung cancer cell line. The selective endonuclease activity of TNA enzymes offers a new route for designing XNA-based molecular tools useful for future biomedical applications.
Jackson et al. , in 2019, reported crystal structures of a natural Bst DNA polymerase that capture the post-translocated product of DNA synthesis on templates composed entirely of 2'-deoxy-2'-fluoro-β-d-arabino nucleic acid (FANA) and α-l-threofuranosyl nucleic acid (TNA). The analysis of the enzyme active site revealed the structural plasticity as a possible mechanism for XNA-dependent DNA synthesis providing insights into the construction of variants with improved activity. Figure 2 shows structural models of the Bst DNA polymerase I TNA/DNA complex.
| 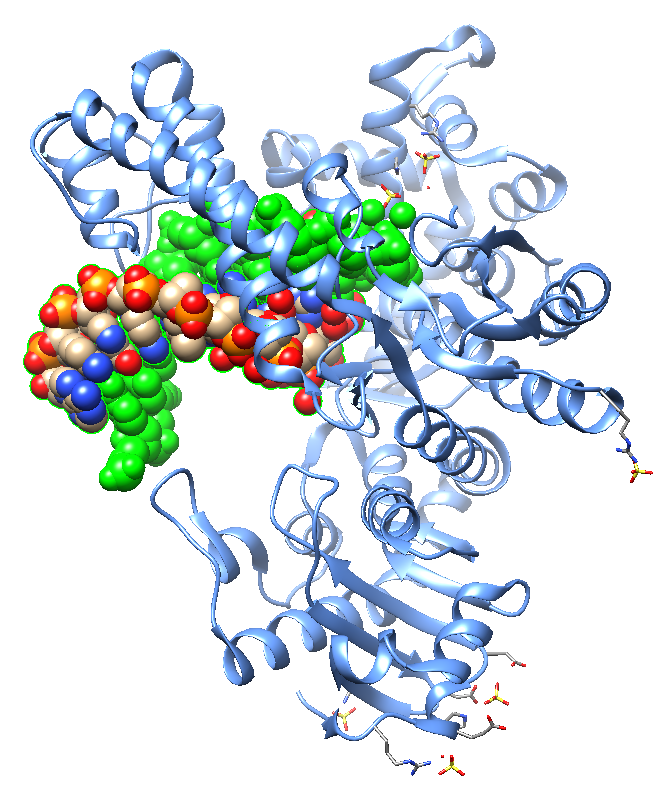 | 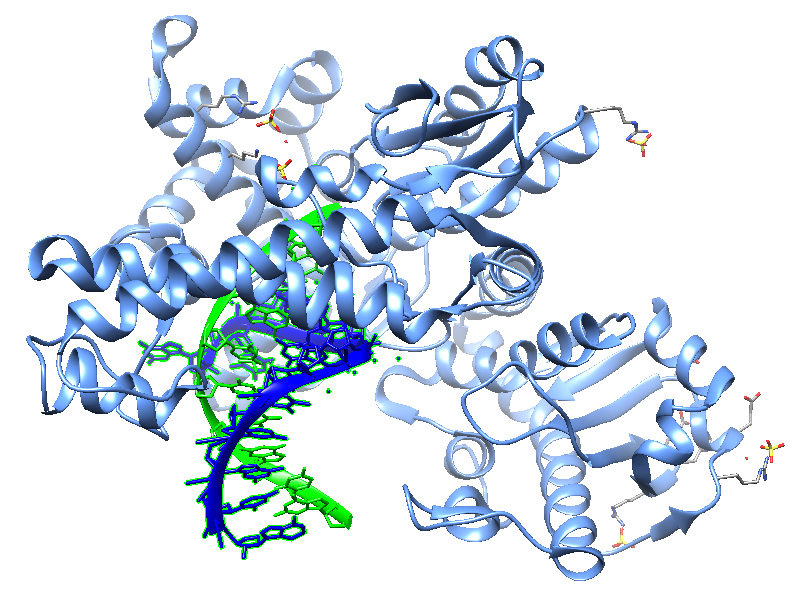 |
| Figure 2: Two views of the crystal structure of the Bst DNA polymerase I TNA/DNA binary complex (PDB ID 6MU5). The DNA oligonucleotide is highlighted in green and the TNA oligonucleotide in blue. This natural DNA polymerase functions as an XNA reverse transcriptase [pdb/6MU5]. |
Reference
Chaput J.C., Ichida,J.K. and Szostak,J.W. (2003); DNA polymerase-mediated DNA synthesis on a TNA template. J. Am. Chem. Soc., 125, 856–857. [PubMed]
Chim, N., Shi, C., Sau, S.P. et al. Structural basis for TNA synthesis by an engineered TNA polymerase. Nat Commun 8, 1810 (2017). [PMC]
Jackson LN, Chim N, Shi C, Chaput JC. Crystal structures of a natural DNA polymerase that functions as an XNA reverse transcriptase. Nucleic Acids Res. 2019 Jul 26;47(13):6973-6983. [PMC; PDB ID 6MU5] Reported crystal structures of natural Bst DNA polymerase that capture the post-translocated product of DNA synthesis on templates composed entirely of 2′-deoxy-2′-fluoro-β-d-arabino nucleic acid (FANA) and α-l-threofuranosyl nucleic acid (TNA).
Kempeneers V, Vastmans K, Rozenski J, Herdewijn P. Recognition of threosyl nucleotides by DNA and RNA polymerases. Nucleic Acids Res. 2003 Nov 1;31(21):6221-6. [PMC]
Lee EM, Setterholm NA, Hajjar M, Barpuzary B, Chaput JC. Stability and mechanism of threose nucleic acid toward acid-mediated degradation. Nucleic Acids Res. 2023 Oct 13;51(18):9542-9551. [PMC] “Lee et al. compared the stability and mechanism of acid-mediated degradation of α-l-threose nucleic acid (TNA) to that of natural DNA and RNA and found that under acidic conditions and elevated temperature (pH 3.3 at 90°C), TNA was significantly more resistant to acid-mediated degradation than DNA and RNA.“
Li Q, Maola VA, Chim N, Hussain J, Lozoya-Colinas A, Chaput JC. Synthesis and Polymerase Recognition of Threose Nucleic Acid Triphosphates Equipped with Diverse Chemical Functionalities. J Am Chem Soc. 2021 Oct 27;143(42):17761-17768. [PubMed; PDB ID 7RSU]
Liao JY, Bala S, Ngor AK, Yik EJ, Chaput JC. P(V) Reagents for the Scalable Synthesis of Natural and Modified Nucleoside Triphosphates. J Am Chem Soc. 2019 Aug 28;141(34):13286-13289. [JACS]
Liu LS, Leung HM, Tam DY, Lo TW, Wong SW, Lo PK. α-l-Threose Nucleic Acids as Biocompatible Antisense Oligonucleotides for Suppressing Gene Expression in Living Cells. ACS Appl Mater Interfaces. 2018 Mar 21;10(11):9736-9743. [PubMed]
Lynnette N Jackson, Nicholas Chim, Changhua Shi, John C Chaput, Crystal structures of a natural DNA polymerase that functions as an XNA reverse transcriptase, Nucleic Acids Research, Volume 47, Issue 13, 26 July 2019, Pages 6973–6983. [NAR]
McCloskey CM, Li Q, Yik EJ, Chim N, Ngor AK, Medina E, Grubisic I, Co Ting Keh L, Poplin R, Chaput JC. Evolution of Functionally Enhanced α-l-Threofuranosyl Nucleic Acid Aptamers. ACS Synth Biol. 2021 Nov 19;10(11):3190-3199. [ACS]
Mei H, Chaput J. Synthesis of a Fluorescent Cytidine TNA Triphosphate Analogue. Methods Mol Biol. 2019;1973:27-37. [PubMed]
Schöning K, Scholz P, Guntha S, Wu X, Krishnamurthy R, Eschenmoser A. Chemical etiology of nucleic acid structure: the alpha-threofuranosyl-(3'-->2') oligonucleotide system. Science. 2000 Nov 17;290(5495):1347-51. [PubMed]
Wang Y, Wang Y, Song D, Sun X, Li Z, Chen JY, Yu H. An RNA-cleaving threose nucleic acid enzyme capable of single point mutation discrimination. Nat Chem. 2022 Mar;14(3):350-359. [PubMed]
---...---
" Bio-Synthesis provides a full spectrum of high quality custom oligonucleotide modification services including back-bone modifications, conjugation to fatty acids, biotinylation by direct solid-phase chemical synthesis or enzyme-assisted approaches to obtain artificially modified oligonucleotides, such as BNA antisense oligonucleotides, mRNAs or siRNAs, containing a natural or modified backbone, as well as base, sugar and internucleotide linkages.
Bio-Synthesis also provides biotinylated mRNA and long circular oligonucleotides".
---...---