Pancreatic cancer is the 4th leading cause of cancer deaths in the U. S. and may claim ~43,000 lives in this year. Typically, it occurs at ages >45, irrespective of gender. Pancreatic cancer is difficult to diagnose due to the lack of specific symptoms, thus the majority of the diagnosed present advanced-stage disease. In early-stage pancreatic cancer, surgical resection may increase 5-year survival rate to ~20% (Aslan et al., 2018). For all-stage pancreatic cancers, 5-year survival rate remains ~7%. Chemotherapy, i.e. 5-flourouracil, nab-paclitaxel (albumin–packaged Taxol), gemcitabine or topoisomerase inhibitor, may increase overall survival moderately (~8.5 months following gemcitabine and nab-paclitaxel treatment), and combining with radiotherapy exacerbates side effects. The efficacy of targeted therapies inhibiting growth signaling pathways (or stroma) and immunotherapies is being explored.
The pancreas located behind the stomach plays a critical role in digestion. Its exocrine cells secrete enzymes to break down lipids, carbohydrates and proteins while endocrine cells release hormones (ex. insulin to regulate sugar level). Nearly 85% of all pancreatic cancers occur in exocrine cells, and pancreatic ductal adenocarcinoma (PDAC) rising in the cells lining the pancreatic ducts represents the most aggressive type. Pancreatic neuroendocrine tumor (PanNET) originating in the endocrine cells represents a minority. Most of pancreatic cancers harbor chromosomal alterations (ex. translocation, inversion, deletion) and KRAS (>95%), p53, CDKN2A and SMAD4 genes are frequently mutated (BRCA1/BRCA2 is mutated in a subset) (Cicenas et al., 2017).
In normal cells, glucose is metabolized (by glycolysis) to pyruvate, which is converted to acetyl-CoA to enter tricarboxylic acid cycle (TCA) and produce NADH to drive oxidative phosphorylation and generate the maximal amount of ATP in mitochondria. In cancer cells, however, this pathway is bypassed in favor of increased glycolysis (‘Warburg effect’ by O. Warburg, Nobel prize 1931) to generate glycolytic intermediates to support their uncontrolled growth (Warburg, 1956). In PDAC cells, constitutively activated signaling by mutant K-RAS oncoprotein facilitates glycolysis by modulating transporters to increase glucose uptake. It also facilitates shunting of the glycolysis intermediates to ‘pentose phosphate pathway’ to generate ribose 5’-phosphate necessary for DNA replication of rapidly dividing cancer cells (Bryant et al., 2014).
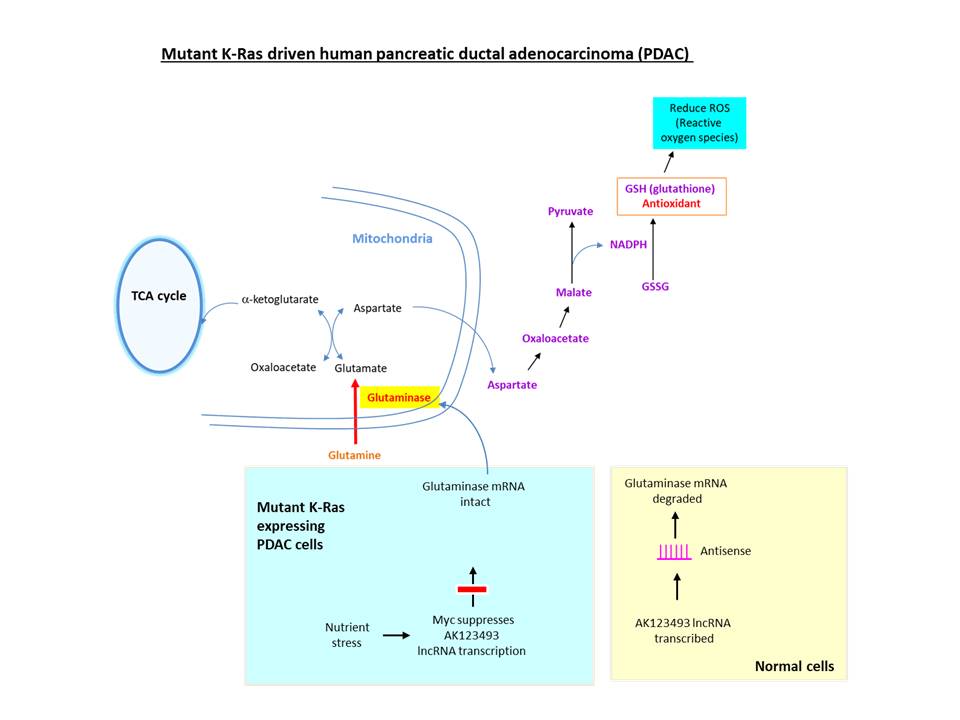
To maintain redox balance, proliferative cells consume greater amounts of glutamine (Eagle, 1955). In K-RAS driven PDAC cells, instead of converting glutamate (derived from glutamine) to a-ketoglutarate (an intermediate of TCA cycle), it is converted to malate, which is metabolized to yield NADPH necessary for generating the antioxidant glutathione. The hydrolysis of glutamine to glutamate is catalyzed by the enzyme glutaminase, which is overexpressed in PDAC (Chakrabarti et al., 2015), breast cancer, prostate cancer, etc.
Long coding RNA (lncRNA) refers to transcripts >200 bp to distinguish from ‘small noncoding RNA’ (ex. siRNA, microRNA, small nucleolar RNA) and >270,000 lncRNA transcripts (Ma et al., 2019) may exist in humans. Most lncRNAs have similar genomic structure (~42% consist of 2 exons) as protein-coding genes, transcribed as independent transcription unit from promoters epigenetically regulated through histone modification, undergo splicing (Derrien et al., 2012), and exhibit reduced/tissue-specific expression. They include intergenic lncRNA, antisense lncRNA, sense lncRNA (located in intron, share exon or overlap exons of protein-coding gene). LncRNA may affect gene transcription directly (as co-factor or via RNA-DNA triplex formation) or indirectly, regulate post-transcriptionally via hybridizing to mRNA to alter splicing/translation, or induce RNA interference for degradation. Consistent with their presence in chromatin, lncRNAs are involved in epigenetically regulating genes during embryogenesis, imprinting, X chromosome inactivation and telomere protection.
To understand the molecular mechanism regulating glutaminase, the investigators at Huazhong University of Science and Technology (China) examined long noncoding RNAs (lncRNAs) dysregulated in PDAC. AK123493 is an antisense lncRNA located in intron 17 of human glutaminase gene, which is underexpressed in PDAC. A ‘co-RNA FISH’ assay revealed that both RNAs are co-localized in the same nuclear foci, indicating that AK123493 lncRNA may hybridize to GLS mRNA (Deng et al., 2019). For the assay, fluorescently labeled RNA probes were generated by incorporating modified UTP (with NH2 group linked to the base) through in vitro transcription, followed by conjugation with amine-reactive fluorescent dyes. This, and other data, suggested that AK123493 lncRNA targets glutaminase mRNA to reduce its expression via inducing RNA interference. Further, they suggested that nutrient stress (caused by glutamine depletion) may activate Myc oncoprotein to inhibit the transcription of AK123493 lncRNA to increase the glutaminase level.
Bio-Synthesis, Inc. specializes in oligonucleotide modification and provides an extensive array of chemically modified nucleoside analogues (over ~200) including bridged nucleic acid (BNA). A number of options are available to label oligonucleotides (DNA or RNA) with fluorophores either terminally or internally as well as conjugate to peptides. It recently acquired a license from BNA Inc. of Osaka, Japan, for the manufacturing and distribution of BNANC, a third generation of BNA oligonucleotides. To meet the demands of therapeutic application, its oligonucleotide products are approaching GMP grade. Bio-Synthesis, Inc. has recently entered into collaborative agreement with Bind Therapeutics, Inc. to synthesize miR-21 blocker using BNA for triple negative breast cancer. The BNA technology that we offer provides superior, unequalled advantages in base stacking, binding affinity, aqueous solubility and nuclease resistance. It also improves the formation of duplexes and triplexes by reducing the repulsion between the negatively charged phosphates of the oligonucleotide backbone. Its single-mismatch discriminating power was especially useful for diagnosis (ex. FISH using DNA probe). More importantly, BNA oligonucleotide exhibits lesser toxicity than other modified nucleotides for clinical application.
https://www.biosyn.com/oligonucleotide-modification-services.aspx
https://www.biosyn.com/tew/Specific-labeling-of-RNA.aspx
https://www.biosyn.com/labeledrnaprobes.aspx#!
References
Aslan M, Shahbazi R, Ulubayram K, Ozpolat B. Targeted Therapies for Pancreatic Cancer and Hurdles Ahead. (2018). Anticancer Res 38:6591-6606. PMID: 30504367 doi: 10.21873/anticanres.13026.
Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. (2014) Trends Biochem Sci. 39:91-100. PMID: 24388967 doi: 10.1016/j.tibs.2013.12.004
Chakrabarti G, Moore ZR, Luo X, Ilcheva M, Ali A, Padanad M, et al. Targeting glutamine metabolism sensitizes pancreatic cancer to PARP-driven metabolic catastrophe induced by ß-lapachone. (2015) Cancer Metab 3:12. PMID: 26462257 doi: 10.1186/s40170-015-0137-1. eCollection 2015.
Cicenas J, Kvederaviciute K, Meskinyte I, Meskinyte-Kausiliene E, Skeberdyte A, Cicenas J. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 Mutations in Pancreatic Cancer. (2017) Cancers (Basel). 9(5). pii: E42. PMID: 28452926 doi: 10.3390/cancers9050042.
Deng SJ, Chen HY, Zeng Z, Deng S, Zhu S, Ye Z, et al. Nutrient Stress-Dysregulated Antisense lncRNA GLS-AS Impairs GLS-Mediated Metabolism and Represses Pancreatic Cancer Progression. (2019) Cancer Res. 79:1398-1412. PMID: 30563888 doi: 10.1158/0008-5472.CAN-18-0419.
Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. (2012) Genome Res 22:1775-89. PMID: 22955988 doi: 10.1101/gr.132159.111.
Eagle H. Nutrition needs of mammalian cells in tissue culture. (1955) Science 122:501-14. PMID: 13255879 DOI: 10.1126/science.122.3168.501
Ma L, Cao J, Liu L, Du Q, Li Z, Zou D, Bajic VB, Zhang Z. LncBook: a curated knowledgebase of human long non-coding RNAs. (2019) Nucleic Acids Res 47:2699. PMID: 30715521 doi: 10.1093/nar/gkz073
Warburg O. On the origin of cancer cells. (1956) Science 123:309-14. PMID: 13298683