The current pandemic is characterized by the intermittent waves of COVID-19 infection. This is due to the high mutation rate of SARS-CoV-2 coronavirus, which is caused, in part, by the low fidelity of the virally encoded RdRP (RNA-dependent RNA polymerase that replicates its genome). A relatively high mutation rate is being maintained despite the proofreading capacity rendered by its Nsp14 gene product. It has led to the evolution of a series of SARS-CoV-2 variants including B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), and B.1.617.2 (Delta).
Following the introduction of vaccines expressing SARS-CoV-2 spike protein in the spring of 2021, a decline in the rate of infection was observed. The reduction is attributed to the induction of humoral (antibody) as well as cellular immunity (cytotoxic T cell) against the virus. However, the vaccine-induced (B cell-based) antibody response has been severely weakened against the currently circulating Omicron variant, which may account for its high rate of infectivity/transmission. Nonetheless, the vaccine-induced T cell-based immunity against Omicron infected cells appears preserved, which may explain the lesser severity/hospitalization observed. Incidentally, natural immunity acquired following the COVID-19 infection provided a comparable level of immunity for the recovered (Guo et al., 2022; Ridgway et al., 2022).
A recent report described that T7 bacteriophage polymerase (lacks proofreading capacity) used to prepare mRNA vaccine (encoding COVID-19 spike protein) exhibits nearly twice the rate of mutation as COVID-19’s RdRP (2.3 x 10-4). Hence, a single mRNA molecule encoding the COVID-19 spike protein (1273 residues) transcribed by T7 polymerase may harbor one nucleotide variant (on average). Consistently, sequencing of the mRNA vaccine (Pfizer-BioNTech) revealed that the mRNAs harbor point mutations (G-to-A being more common than C-to-T; 67% of point mutations being nonsynonymous, i.e. changes amino acids) along with other types of mutations (ex. Insertion, deletion). Thus, the COVID-19 spike protein expressed by the mRNA vaccine appears to have been heterogeneous (Herman et al., 2022).
To address these concerns, nonvaccine-based therapeutics are increasingly sought. Also, the availability of the latter may help immunocompromised individuals, who are unable to mount immunological response despite the vaccination. For cancer patients, CDC (Centers for Disease Control and Prevention, United States) recommends COVID-19 vaccination at periods devoid of treatments that may suppress immunity, i.e. chemotherapy, radiotherapy, stem cell transplantation therapy. (Clinical Guidance for COVID-19 Vaccination | CDC ). However, for patients undergoing immunotherapy (Immune Checkpoint Inhibitor treatment designed to boost immunity against tumor), the potential to increase adverse side effects associated with mRNA vaccine for COVID-19 is being explored (Brest et al., 2022).
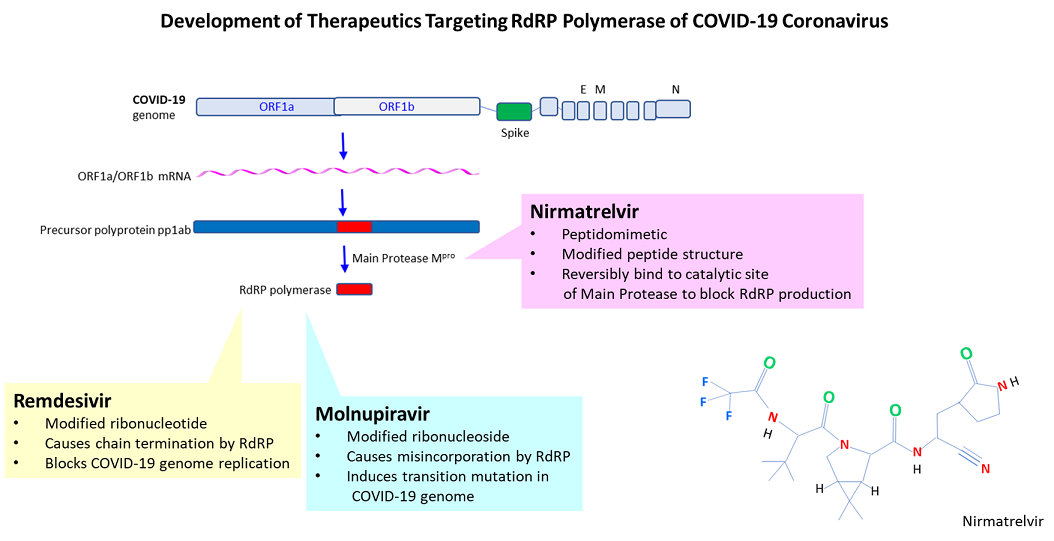
Among the current anti-COVID-19 drugs approved by FDA under EUA (Emergency Use Authorization) is the prodrug Remdesivir (antiviral drug for hepatitis C; Gilead Sciences), which undergoes several reactions to become activated. The triphosphate form of the modified ribonucleotide GS-441524 (Gilead Sciences) causes chain termination, blocking genome replication by RdRP (Beigel et al., 2020).
Another pharmacological approach seeks to inactivate Covid-19 coronavirus by introducing errors to its genome. After administering, the prodrug Molnupiravir (antiviral drug for influenza; Merck), β-d-N4 -hydroxycytidine, is converted to its active triphosphate form. RdRP polymerase misincorporates Molnupiravir in place of cytidine, which causes (depending on its tautomeric status) the incorporation of ATP (instead of the normal GTP) in the complementary strand, resulting in the G-to-A transition. Because of the concern that the drug may mutate COVID-19 into a more virulent variant, it was narrowly approved by FDA (for individuals who lack other treatment options) (Malone et al., 2021).
The 3rd FDA approved drug under EUA is Paxlovid (for high-risk patients), which is comprised of dual drugs (nirmatrelvir and ritonavir). Ritonavir functions to elevate the blood level of nirmatrelvir by suppressing its degradation (by cytochrome p450) (Hossain et al., 2017). Nirmatrelvir inhibits the ‘main protease’ (Mpro), a cysteine protease that cleaves the precursor polyprotein pp1ab of COVID-19 into 16 nonstructural proteins including RdRP. In 2005, Yang and colleagues reported the isolation of the wide-spectrum inhibitor ‘N3’, a peptidomimetic that forms an irreversible covalent bond with the cysteine residue in the catalytic site (Yang et al, 2005; Jin et al., 2020).
In 2020, Hoffman and colleagues (Pfizer and Southern Research Institute, United States) reported the isolation of the potent inhibitor ‘PF-00835231’ of main protease (Hoffman et al., 2020). The latter peptidomimetic was further modified for oral uptake (PF-07321332, Nirmatrelvir; IC50 = 4 nM), whose nitrile group forms a reversible covalent bond with the cysteine’s thiol group (Zhao et al., 2021; Duveau et al., 2022; Owen et al., 2021). Phase II/III clinical trial conducted on unvaccinated, symptomatic high-risk patients reported that treatment with nirmatrelvir and ritonavir within 5 days after the onset of symptoms reduces hospitalization (6.2 to 0.7%) or deaths (1.1 to 0%) (Hammod et al., 2022).
Despite these promising results, several reports documented specific mutation(s) in main protease that confers resistance to nirmatrelvir. One report described that the changes in the residues (L50F, E166A, and L167F) of main protease give rise to nirmatrelvir-resistant COVID-19 coronavirus (Jochmans et al., 2022). Another report described similar findings with the L50F or E166A mutation (also reduced the catalytic activity of Mpro) (Zhou et al., 2022). Further, it was suggested that such mutations might already be extant in the circulating COVID-19 virus population, which may become prevalent with wider use of the drug (Sedova et al., 2022). Of relevance, a mutation that confers resistance to remdesivir has already been identified in patients (Gandhi et al., 2022).
The key to preventing an epidemic is the ability to diagnose the infected early to preempt further propagation. For this, Bio-Synthesis, Inc. provides primers and probes (as well as synthetic RNA control) for COVID-19 diagnosis via RT-PCR assay. It specializes in oligonucleotide modification and provides an extensive array of chemically modified nucleoside analogs (over ~200) including bridged nucleic acid (BNA) in addition to mRNA synthesis. A number of options are available to label oligonucleotides (DNA or RNA) with fluorophores either terminally or internally as well as to conjugate to peptides or antibodies. It provides custom conjugation of small molecules such as chemical drugs, metabolites and labeled compounds with synthetic or natural polymers (enzymes, peptide, protein, oligonucleotide, antibody, dendrimer, nanoparticle, etc). It recently acquired a license from BNA Inc. of Osaka, Japan, for the manufacturing and distribution of BNANC, the third generation of BNA oligonucleotides. To meet the demands of therapeutic application, its oligonucleotide products are approaching GMP grade. It has recently entered into collaborative agreement with Bind Therapeutics, Inc. to synthesize miR-21 blocker using BNA for triple-negative breast cancer. The BNA technology provides superior, unequaled advantages in base stacking, binding affinity, aqueous solubility and nuclease resistance. It also improves the formation of duplexes and triplexes by reducing the repulsion between the negatively charged phosphates of the oligonucleotide backbone. Its single-mismatch discriminating power is especially useful for diagnosis (ex. FISH using DNA probe). For clinical application, BNA oligonucleotide exhibits lesser toxicity than other modified nucleotides. For therapeutic consideration, peptide modifications may include labeling, conjugation, cyclization, incorporation of unusual amino acids, and modification of side chain and backbone.
https://www.biosyn.com/oligo-flourescent-labeling.aspx
https://www.biosyn.com/tew/Speed-up-Identification-of-COVID19.aspx
https://www.biosyn.com/covid-19.aspx
https://www.biosyn.com/mrna.aspx
https://www.biosyn.com/bioconjugation.aspx
https://www.biosyn.com/tew/Design-Guidelines-for-BNA-based-Oligonucleotide-Probes.aspx#!
https://www.biosyn.com/tew/Remdesivir-and-COVID-19.aspx
Peptide Modifications, Modified Peptide Synthesis - Bio-Synthesis (biosyn.com)
References
Beigel JH, Lane HC, et al. ACTT-1 Study Group Members. Remdesivir for the Treatment of Covid-19 - Final Report. N Engl J Med. 383:1813-1826 (2020). PMID: 32445440
Breast P, Milano G, et al. COVID-19 vaccination and cancer immunotherapy: should they stick together? Br J Cancer 126, 1-3 (2022). PMID: 34799696
Duveau DY, Thomas CJ. The Remarkable Selectivity of Nirmatrelvir. ACS Pharmacol Transl Sci. 5:445-447 (2022). PMID: 35702394
Gandhi S, Ko A, et al., De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: A case report. medRxiv (2021) (Preprint).
Guo L, Wang J, et al. Assessment of Antibody and T-Cell Responses to the SARS-CoV-2 Virus and Omicron Variant in Unvaccinated Individuals Recovered From COVID-19 Infection in Wuhan, China. JAMA Netw Open. 5:e229199 (2022). PMID: 35476069
Hammond J, Rusnak JM; EPIC-HR Investigators, et al. Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with Covid-19. N Engl J Med. 386:1397-1408 (2022). PMID: 35172054
Herman C, Ronca S, et al. RNA polymerase inaccuracy underlies SARS-CoV-2 variants and vaccine heterogeneity. Res Sq. rs.3.rs-1690086 (2022). PMID: 35677076
Hoffman RL, Taggart B, et al. Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19. J Med Chem. 63:12725-12747 (2020. PMID: 33054210
Hossain MA, Greenblatt DJ, et al. Inhibition of human cytochromes P450 in vitro by ritonavir and cobicistat. J Pharm Pharmacol. 69:1786-1793 (2017). PMID: 28960344
Jin Z, Yang H, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 582:289-293 (2020). PMID: 32272481
Jochman D, Vandyck K, et al. The substitutions L50F, E166A and L167F in SARSCoV-2 3CLpro are selected by a protease inhibitor in vitro and confer resistance to nirmatrelvir. bioRxiv (2022) (preprint).
Malone B, Campbell EA. Molnupiravir: coding for catastrophe. Nat Struct Mol Biol. 28:706-708 (2021). PMID: 34518697
Owen DR, Zhu Y, et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 374:1586-1593 (2021). PMID: 34726479
Ridgway JP, Robicsek A, et al. Rates of COVID-19 Among Unvaccinated Adults With Prior COVID-19.
JAMA Netw Open. 5:e227650 (2022). PMID: 35442459
Sedova M, Godzik A, et al., Monitoring for SARS-CoV-2 drug resistance mutations in broad viral populations. bioRxiv (2022). (Preprint)
Yang H, Rao Z, et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 3:e324 (2005). PMID: 16128623
Zhao Y, , Yang H, et al. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell. 13:689-693 (2022). PMID: 34687004
Zhou Y, Gottwein JM, et al. Nirmatrelvir Resistant SARS-CoV-2 Variants with High Fitness in Vitro. bioRxiv (2022) (Preprint )