In recent years, the method of high throughput screening has become a mainstay in both biological and pharmaceutical researches. The ability to identify protein(s) interacting with a target protein through screening an appropriate library greatly expedites the research. By doing so, one may bypass conventional steps entailing biochemical purification plus characterization and/or proteomic analyses.
For pharmaceutical research, high throughput screening has been used to identify small molecule drug-like chemical compounds for treating various disorders. Hence, to suppress the cancer-promoting property of an oncogene-encoded receptor, identifying chemical compounds capable of inhibiting specific receptor function (ex. tyrosine kinase) may be sufficient for pharmacological management. Alternatively, identifying chemical compounds capable of inhibiting the catalytic activity of an enzyme in a biosynthetic pathway may satisfy the pharmacological objective.
Yet, the utility of screening chemical libraries has become increasingly limited with the expanding knowledge concerning the complex biochemical processes underlying human disorders. Recent research has revealed that the downstream signaling step culminating in transcriptional regulation of target genes involves complex interaction by numerous distinct proteins. For instance, the transcriptional regulation by estrogen receptor at the estrogen-responsive elements requires the assembly of 1 to 2 megadalton-sized complex comprised of numerous transcription factors including AP2γ, RARα/γ, GATA3, STAT1, FoxA and AP1 (Liu et al., 2014). In the case of proteosome (~750 kilodalton) involved in protein degradation, the assembly of a functional complex requires the association of a number of distinct subunits and regulatory proteins (Tanaka, 2009).
The research has further uncovered that a significant fraction of protein-to-protein interactions (~40%) are mediated through short peptides. Though small drug-like chemicals have been effective in targeting buried hydrophobic binding pockets like the ligand-binding site of a receptor, peptides are better suited for disrupting protein-to-protein interactions that encompass a greater surface area. Though antibodies also exhibit specificity, they suffer from poor penetration of tissues, instability (easy to denature), membrane impermeability, costly and time-consuming preparation, immunogenicity, etc. This has inspired a greater focus by the pharmaceutical industries on improving various facets of peptides for clinical application, i.e. terminal protection, D-enantiomers, peptide cyclization, conjugation with functional groups (to improve specificity, tissue infiltration, resistance to degradation, etc.) (Lee et al., 2019). Through these modifications, peptides exhibiting 5 times higher binding affinity than chemical compounds have been developed.
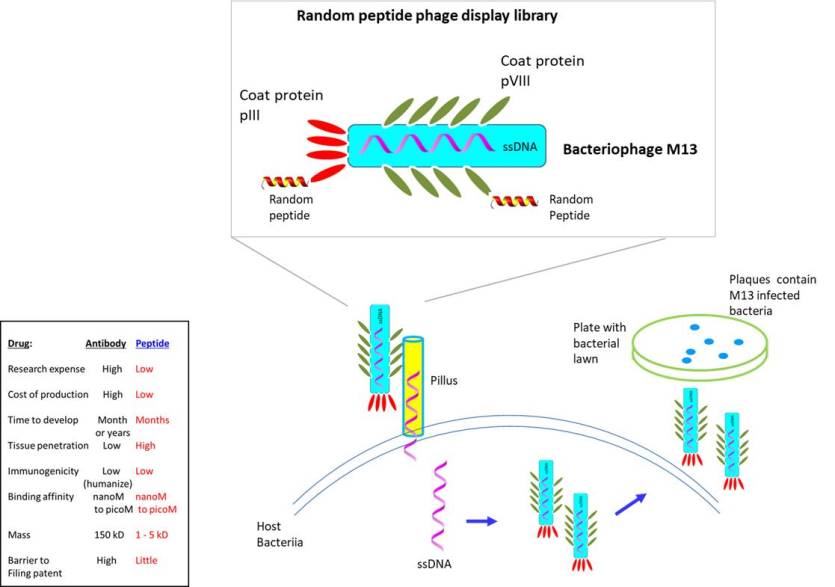
It follows that the ability to identify therapeutic peptides through a high throughput screening of a random peptide library would be highly beneficial. To facilitate such endeavor, a biopanning system that screens random peptide library displayed on the surface of bacteriophage (bacteria infecting virus) was designed by G. Smith (Nobel prize, 2018). The library is based on filamentous bacteriophage (ex. M13, fd, f1) containing single stranded DNA as its genome (Saw et al., 2019). The filament-like shape helps to align and transfer its genetic material through the pilus into the cytoplasm of the host bacteria. To construct the library, random peptides were expressed as part of the minor coat protein pIII or major coat protein pVIII. Since its introduction, phage display library has been used extensively for multiple applications--for ex. to identify novel tumor antigens, enzyme inhibitors, antagonist/agonist of receptor, delivery vector (targeting blood vessels, tissues, cancer) or for other industrial uses.
The key to preventing epidemic is the ability to diagnose the infected early to preempt further propagation. For this, Bio-Synthesis, Inc. provides primers and probes (as well as synthetic RNA control) for COVID-19 diagnosis via RT-PCR assay. It specializes in oligonucleotide modification and provides an extensive array of chemically modified nucleoside analogues (over ~200) including bridged nucleic acid (BNA). A number of options are available to label oligonucleotides (DNA or RNA) with fluorophores either terminally or internally as well as to conjugate to peptides or antibodies. It recently acquired a license from BNA Inc. of Osaka, Japan, for the manufacturing and distribution of BNANC, a third generation of BNA oligonucleotides. To meet the demands of therapeutic application, its oligonucleotide products are approaching GMP grade. Bio-Synthesis, Inc. has recently entered into collaborative agreement with Bind Therapeutics, Inc. to synthesize miR-21 blocker using BNA for triple negative breast cancer. The BNA technology provides superior, unequalled advantages in base stacking, binding affinity, aqueous solubility and nuclease resistance. It also improves the formation of duplexes and triplexes by reducing the repulsion between the negatively charged phosphates of the oligonucleotide backbone. Its single-mismatch discriminating power is especially useful for diagnosis (ex. FISH using DNA probe). For clinical application, BNA oligonucleotide exhibits lesser toxicity than other modified nucleotides.
https://www.biosyn.com/oligo-flourescent-labeling.aspx
https://www.biosyn.com/tew/Speed-up-Identification-of-COVID19.aspx
https://www.biosyn.com/covid-19.aspx
https://www.biosyn.com/peptide-synthesis.aspx
References
Lee AC, Harris JL, et al. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int J Mol Sci. 20:2383 (2019). PMID: 31091705
Liu Z, Merkurjev D, et al. Enhancer activation requires trans-recruitment of a mega transcription factor complex. Cell 159:358-73 (2014). PMID: 25303530
Saw PE, Song EW. Phage display screening of therapeutic peptide for cancer targeting and therapy. Protein Cell 10:787-807 (2019). PMID: 31140150
Murata S, Yashiroda H, et al. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 10:104-15 (2009). PMID: 19165213